Reconstruction of gene regulatory networks for Caenorhabditis elegans using tree-shaped gene expression data
- PMID: 39133097
- PMCID: PMC11318059
- DOI: 10.1093/bib/bbae396
Reconstruction of gene regulatory networks for Caenorhabditis elegans using tree-shaped gene expression data
Abstract
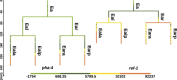
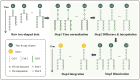
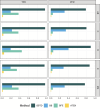
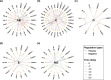
Constructing gene regulatory networks is a widely adopted approach for investigating gene regulation, offering diverse applications in biology and medicine. A great deal of research focuses on using time series data or single-cell RNA-sequencing data to infer gene regulatory networks. However, such gene expression data lack either cellular or temporal information. Fortunately, the advent of time-lapse confocal laser microscopy enables biologists to obtain tree-shaped gene expression data of Caenorhabditis elegans, achieving both cellular and temporal resolution. Although such tree-shaped data provide abundant knowledge, they pose challenges like non-pairwise time series, laying the inaccuracy of downstream analysis. To address this issue, a comprehensive framework for data integration and a novel Bayesian approach based on Boolean network with time delay are proposed. The pre-screening process and Markov Chain Monte Carlo algorithm are applied to obtain the parameter estimates. Simulation studies show that our method outperforms existing Boolean network inference algorithms. Leveraging the proposed approach, gene regulatory networks for five subtrees are reconstructed based on the real tree-shaped datatsets of Caenorhabditis elegans, where some gene regulatory relationships confirmed in previous genetic studies are recovered. Also, heterogeneity of regulatory relationships in different cell lineage subtrees is detected. Furthermore, the exploration of potential gene regulatory relationships that bear importance in human diseases is undertaken. All source code is available at the GitHub repository https://github.com/edawu11/BBTD.git.
Keywords: Bayesian statistics; Boolean network; data integration; gene regulatory network; tree-shaped gene expression data.
© The Author(s) 2024. Published by Oxford University Press.
Figures

Similar articles
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
A full bayesian approach for boolean genetic network inference.PLoS One. 2014 Dec 31;9(12):e115806. doi: 10.1371/journal.pone.0115806. eCollection 2014. PLoS One. 2014. PMID: 25551820 Free PMC article.
-
Inference of cellular level signaling networks using single-cell gene expression data in Caenorhabditis elegans reveals mechanisms of cell fate specification.Bioinformatics. 2017 May 15;33(10):1528-1535. doi: 10.1093/bioinformatics/btw796. Bioinformatics. 2017. PMID: 28011782
-
A review on the computational approaches for gene regulatory network construction.Comput Biol Med. 2014 May;48:55-65. doi: 10.1016/j.compbiomed.2014.02.011. Epub 2014 Feb 24. Comput Biol Med. 2014. PMID: 24637147 Review.
-
Learning Differential Module Networks Across Multiple Experimental Conditions.Methods Mol Biol. 2019;1883:303-321. doi: 10.1007/978-1-4939-8882-2_13. Methods Mol Biol. 2019. PMID: 30547406 Review.
References
-
- Akers K, Murali T. Gene regulatory network inference in single-cell biology. Curr Opin Syst Biol 2021; 26:87–97. 10.1016/j.coisb.2021.04.007. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

