Histone H4 acetylation differentially modulates proliferation in adult oligodendrocyte progenitors
- PMID: 39133301
- PMCID: PMC11318668
- DOI: 10.1083/jcb.202308064
Histone H4 acetylation differentially modulates proliferation in adult oligodendrocyte progenitors
Abstract
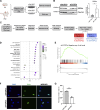
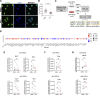
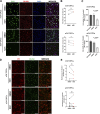
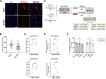
Adult oligodendrocyte progenitors (aOPCs) generate myelinating oligodendrocytes like neonatal progenitors (nOPCs), and they also display unique functional features. Here, using unbiased histone proteomics analysis and ChIP sequencing analysis of PDGFRα+ OPCs sorted from neonatal and adult Pdgfra-H2B-EGFP reporter mice, we identify the activating H4K8ac histone mark as enriched in the aOPCs. We detect increased occupancy of the H4K8ac activating mark at chromatin locations corresponding to genes related to the progenitor state (e.g., Hes5, Gpr17), metabolic processes (e.g., Txnip, Ptdgs), and myelin components (e.g., Cnp, Mog). aOPCs showed higher levels of transcripts related to lipid metabolism and myelin, and lower levels of transcripts related to cell cycle and proliferation compared with nOPCs. In addition, pharmacological inhibition of histone acetylation decreased the expression of the H4K8ac target genes in aOPCs and decreased their proliferation. Overall, this study identifies acetylation of the histone H4K8 as a regulator of the proliferative capacity of aOPCs.
© 2024 Dansu et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
-
- Balasubramanyam, K., Altaf M., Varier R.A., Swaminathan V., Ravindran A., Sadhale P.P., and Kundu T.K.. 2004. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 279:33716–33726. 10.1074/jbc.M402839200 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

