Preclinical spheroid models identify BMX as a therapeutic target for metastatic MYCN nonamplified neuroblastoma
- PMID: 39133652
- PMCID: PMC11383371
- DOI: 10.1172/jci.insight.169647
Preclinical spheroid models identify BMX as a therapeutic target for metastatic MYCN nonamplified neuroblastoma
Abstract
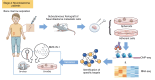
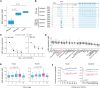
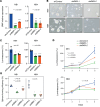
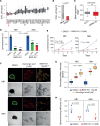
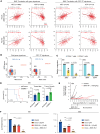
The development of targeted therapies offers new hope for patients affected by incurable cancer. However, multiple challenges persist, notably in controlling tumor cell plasticity in patients with refractory and metastatic illness. Neuroblastoma (NB) is an aggressive pediatric malignancy originating from defective differentiation of neural crest-derived progenitors with oncogenic activity due to genetic and epigenetic alterations and remains a clinical challenge for high-risk patients. To identify critical genes driving NB aggressiveness, we performed combined chromatin and transcriptome analyses on matched patient-derived xenografts (PDXs), spheroids, and differentiated adherent cultures derived from metastatic MYCN nonamplified tumors. Bone marrow kinase on chromosome X (BMX) was identified among the most differentially regulated genes in PDXs and spheroids versus adherent models. BMX expression correlated with high tumor stage and poor patient survival and was crucial to the maintenance of the self-renewal and tumorigenic potential of NB spheroids. Moreover, BMX expression positively correlated with the mesenchymal NB cell phenotype, previously associated with increased chemoresistance. Finally, BMX inhibitors readily reversed this cellular state, increased the sensitivity of NB spheroids toward chemotherapy, and partially reduced tumor growth in a preclinical NB model. Altogether, our study identifies BMX as a promising innovative therapeutic target for patients with high-risk MYCN nonamplified NB.
Keywords: Cancer; Epigenetics; Oncogenes; Therapeutics.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

