Unbalanced MYC break-apart FISH patterns indicate the presence of a MYC rearrangement in HGBCL-DH-BCL2
- PMID: 39133931
- PMCID: PMC11487639
- DOI: 10.1182/blood.2024025603
Unbalanced MYC break-apart FISH patterns indicate the presence of a MYC rearrangement in HGBCL-DH-BCL2
Abstract
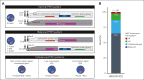
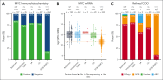
Fluorescence in situ hybridization (FISH) using break-apart probes is recommended for identifying high-grade B-cell lymphoma with MYC and BCL2 rearrangements (HGBCL-DH-BCL2). Unbalanced MYC break-apart patterns, in which the red or green signal is lost, are commonly reported as an equivocal result by clinical laboratories. In a cohort of 297 HGBCL-DH-BCL2, 13% of tumors had unbalanced MYC break-apart patterns with loss of red (LR; 2%) or loss of green (LG; 11%) signal. To determine the significance of these patterns, MYC rearrangements were characterized by sequencing in 130 HGBCL-DH-BCL2, including 3 LR and 14 LG tumors. A MYC rearrangement was identified for 71% of tumors with LR or LG patterns, with the majority involving immunoglobulin loci or other recurrent MYC rearrangement partners. The architecture of these rearrangements consistently preserved the rearranged MYC allele, with the MYC gene predicted to be on the derivative chromosome containing the signal that is still present in nearly all cases. MYC protein expression, MYC messenger RNA expression, and the proportion of tumors expressing the dark-zone signature was not significantly different between balanced and unbalanced groups. These results support a recommendation that unbalanced MYC break-apart FISH patterns be reported as positive for MYC rearrangement in the context of diagnosing HGBCL-DH-BCL2.
© 2024 American Society of Hematology. Published by Elsevier Inc. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
Conflict of interest statement
Conflict-of-interest disclosure: T.T. has received honoraria from AstraZeneca and Merck and research funding from Pfizer. J.W.C. has provided consultancy/expert testimony for Bayer and received honoraria from BeiGene. D.V. reports consulting/honoraria from AbbVie, AstraZeneca, BeiGene, Bristol Myers Squibb (BMS)/Celgene, Gilead/Kite, Incyte, Janssen, Merck, Roche, and ZetaGene and research funding (to the institution) from AstraZeneca and Roche. A.S.G. reports consultancy/honoraria from AbbVie, AstraZeneca, BeiGene, and Janssen and research funding (to the institution) from AbbVie, AstraZeneca, Janssen, and Roche. L.H.S. reports consulting/honoraria from AbbVie, Amgen, AstraZeneca, BeiGene, BMS/Celgene, Genmab, Kite/Gilead, Incyte, Janssen, Merck, Seagen, and Roche/Genentech and research funding from Roche/Genentech and Teva. K.J.S. reports honoraria/consulting fees from BMS, Merck, Seagen, and AbbVie; research funding from BMS; and has served on the drug safety monitoring committee for Regeneron. C.S. has performed consultancy for Bayer and Eisai and has received research funding from Epizyme and Trillium Therapeutics. D.W.S. has received honoraria from AbbVie, AstraZeneca, Genmab, Roche, and Veracyte; research funding from Roche/Genentech; and is an inventor on patents describing the use of gene expression to subtype aggressive B-cell lymphomas, one of which is licensed to NanoString Technologies. The remaining authors declare no competing financial interests.
Figures


Comment in
-
FISHing for clarity in double-hit lymphomas.Blood. 2024 Oct 10;144(15):1550-1552. doi: 10.1182/blood.2024026379. Blood. 2024. PMID: 39388163 No abstract available.
References
-
- Alduaij W, Collinge B, Ben-Neriah S, et al. Molecular determinants of clinical outcomes in a real-world diffuse large B-cell lymphoma population. Blood. 2023;141(20):2493–2507. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

