HMGA1 orchestrates chromatin compartmentalization and sequesters genes into 3D networks coordinating senescence heterogeneity
- PMID: 39134516
- PMCID: PMC11319441
- DOI: 10.1038/s41467-024-51153-8
HMGA1 orchestrates chromatin compartmentalization and sequesters genes into 3D networks coordinating senescence heterogeneity
Abstract
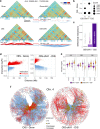
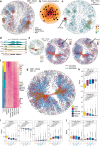
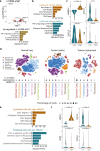
HMGA1 is an abundant non-histone chromatin protein that has been implicated in embryonic development, cancer, and cellular senescence, but its specific role remains elusive. Here, we combine functional genomics approaches with graph theory to investigate how HMGA1 genomic deposition controls high-order chromatin networks in an oncogene-induced senescence model. While the direct role of HMGA1 in gene activation has been described previously, we find little evidence to support this. Instead, we show that the heterogeneous linear distribution of HMGA1 drives a specific 3D chromatin organization. HMGA1-dense loci form highly interactive networks, similar to, but independent of, constitutive heterochromatic loci. This, coupled with the exclusion of HMGA1-poor chromatin regions, leads to coordinated gene regulation through the repositioning of genes. In the absence of HMGA1, the whole process is largely reversed, but many regulatory interactions also emerge, amplifying the inflammatory senescence-associated secretory phenotype. Such HMGA1-mediated fine-tuning of gene expression contributes to the heterogeneous nature of senescence at the single-cell level. A similar 'buffer' effect of HMGA1 on inflammatory signalling is also detected in lung cancer cells. Our study reveals a mechanism through which HMGA1 modulates chromatin compartmentalization and gene regulation in senescence and beyond.
© 2024. The Author(s).
Conflict of interest statement
A.J.P. is an employee of Altos Labs. P.F. and S.S. are co-founders and shareholders of Enhanc3D Genomics Ltd. All other authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

