Variant-proof high affinity ACE2 antagonist limits SARS-CoV-2 replication in upper and lower airways
- PMID: 39134521
- PMCID: PMC11319446
- DOI: 10.1038/s41467-024-51046-w
Variant-proof high affinity ACE2 antagonist limits SARS-CoV-2 replication in upper and lower airways
Abstract
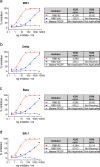
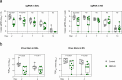
SARS-CoV-2 has the capacity to evolve mutations that escape vaccine- and infection-acquired immunity and antiviral drugs. A variant-agnostic therapeutic agent that protects against severe disease without putting selective pressure on the virus would thus be a valuable biomedical tool that would maintain its efficacy despite the ongoing emergence of new variants. Here, we challenge male rhesus macaques with SARS-CoV-2 Delta-the most pathogenic variant in a highly susceptible animal model. At the time of challenge, we also treat the macaques with aerosolized RBD-62, a protein developed through multiple rounds of in vitro evolution of SARS-CoV-2 RBD to acquire 1000-fold enhanced ACE2 binding affinity. RBD-62 treatment equivalently suppresses virus replication in both upper and lower airways, a phenomenon not previously observed with clinically approved vaccines. Importantly, RBD-62 does not block the development of virus-specific T- and B-cell responses and does not elicit anti-drug immunity. These data provide proof-of-concept that RBD-62 can prevent severe disease from a highly virulent variant.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare the following competing interests: M.S.S. serves on the scientific board of advisors for Moderna and Ocugen. D.C.D. is an inventor on US Patent Application No. 63/147,419 entitled “Antibodies Targeting the Spike Protein of Coronaviruses”. A.V.R., L.P., A.D., A.C., M.G.L. and H.A. are employees of Bioqual. The other authors declare no competing interests.
Figures

Update of
-
RBD-based high affinity ACE2 antagonist limits SARS-CoV-2 replication in upper and lower airways.bioRxiv [Preprint]. 2023 Jun 12:2023.06.09.544432. doi: 10.1101/2023.06.09.544432. bioRxiv. 2023. Update in: Nat Commun. 2024 Aug 12;15(1):6894. doi: 10.1038/s41467-024-51046-w. PMID: 37503026 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

