Defective mitochondrial COX1 translation due to loss of COX14 function triggers ROS-induced inflammation in mouse liver
- PMID: 39134548
- PMCID: PMC11319346
- DOI: 10.1038/s41467-024-51109-y
Defective mitochondrial COX1 translation due to loss of COX14 function triggers ROS-induced inflammation in mouse liver
Abstract
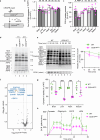
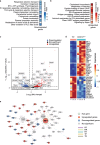
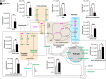
Mitochondrial oxidative phosphorylation (OXPHOS) fuels cellular ATP demands. OXPHOS defects lead to severe human disorders with unexplained tissue specific pathologies. Mitochondrial gene expression is essential for OXPHOS biogenesis since core subunits of the complexes are mitochondrial-encoded. COX14 is required for translation of COX1, the central mitochondrial-encoded subunit of complex IV. Here we describe a COX14 mutant mouse corresponding to a patient with complex IV deficiency. COX14M19I mice display broad tissue-specific pathologies. A hallmark phenotype is severe liver inflammation linked to release of mitochondrial RNA into the cytosol sensed by RIG-1 pathway. We find that mitochondrial RNA release is triggered by increased reactive oxygen species production in the deficiency of complex IV. Additionally, we describe a COA3Y72C mouse, affected in an assembly factor that cooperates with COX14 in early COX1 biogenesis, which displays a similar yet milder inflammatory phenotype. Our study provides insight into a link between defective mitochondrial gene expression and tissue-specific inflammation.
© 2024. The Author(s).
Conflict of interest statement
A.C. is now an employee of Dewpoint Therapeutics GmbH. C.L. is now an employee of BioNTech. The other authors declare no competing interests.
Figures




References
-
- Homberg, B., Rehling, P. & Cruz-Zaragoza, L. D. The multifaceted mitochondrial OXA insertase. Trends Cell Biol. 33, 765–772 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

