Somatic mutational landscape across Indian breast cancer cases by whole exome sequencing
- PMID: 39134585
- PMCID: PMC11319672
- DOI: 10.1038/s41598-024-65148-4
Somatic mutational landscape across Indian breast cancer cases by whole exome sequencing
Abstract
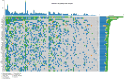
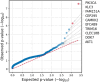
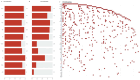
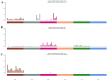
Breast cancer (BC) has emerged as the most common malignancy among females. The genomic profile of BC is diverse in nature and complex due to heterogeneity among various geographically different ethnic groups. The primary objective of this study was to carry out a comprehensive mutational analysis of Indian BC cases by performing whole exome sequencing. The cohort included patients with a median age of 48 years. TTN, TP53, MUC16, SYNE1, and OBSCN were the frequently altered genes found in our cohort. The PIK3CA and KLC3 genes are driver genes implicated in various cellular functions and cargo transportation through microtubules, respectively. Except for CCDC168 and PIK3CA, several gene pairings were found to be significantly linked with co-occurrence. Irrespective of their hormonal receptor status, RTK/RAS was observed with frequently altered signaling pathways. Further analysis of the mutational signature revealed that SBS13, SBS6, and SBS29 were mainly observed in our cohort. This study supplements the discovery of diagnostic biomarkers and provides new therapeutic options for the improved management of BC.
Keywords: Breast cancer; Driver gene; Oncogenic pathway; SBS signature; Whole-exome sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

