Antibiotic resistance gene dynamics in the commensal infant gut microbiome over the first year of life
- PMID: 39134593
- PMCID: PMC11319485
- DOI: 10.1038/s41598-024-69275-w
Antibiotic resistance gene dynamics in the commensal infant gut microbiome over the first year of life
Abstract
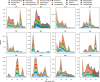
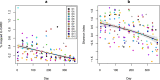
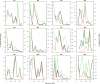
Colonization of the infant gut is an important developmental process characterized by high carriage of antimicrobial resistance genes (ARGs) and high abundances of pathobionts. The horizontal transfer of ARGs to pathogenic bacteria represents a major public health concern. However, there is still a paucity of longitudinal studies surveilling ARGs in healthy infant guts at high temporal resolution. Furthermore, we do not yet have a clear view of how temporal variation in ARG carriage relates to the dynamics of specific bacterial populations, as well as community virulence potential. Here, we performed deep shotgun metagenomic sequencing of monthly fecal samples from a cohort of 12 infants, covering the first year of life to interrogate the infant gut microbiome for ARG content. We further relate ARG dynamics to the dynamics of taxa, virulence potential, as well as the potential for ARG mobilization. We identify a core resistome dominated by efflux systems typically associated with Enterobacteriaceae. Overall ARG carriage declined over the first year of life and showed strong contemporaneous correlation with the population dynamics of Proteobacteria. Furthermore, the majority of ARGs could be further mapped to metagenome-assembled genomes (MAGs) classified to this phylum. We were able to assign a large number of ARGs to E. coli by correlating the temporal dynamics of individual genes with species dynamics, and we show that the temporal dynamics of ARGs and virulence factors are highly correlated, suggesting close taxonomic associations between these two gene classes. Finally, we identify ARGs linked with various categories of mobile genetic elements, demonstrating preferential linkage among mobility categories and resistance to different drug classes. While individual variation in ARG carriage is substantial during infancy there is a clear reduction over the first year of life. With few exceptions, ARG abundances closely track the dynamics of pathobionts and community virulence potential. These findings emphasize the potential for development of resistant pathogens in the developing infant gut, and the importance of effective surveillance in order to detect such events.
Keywords: Antibiotic resistance genes; Gut; Infant; Metagenome; Microbiome; Mobile genetic elements; Temporal dynamics; Virulence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

