Substrates mimicking the blastocyst geometry revert pluripotent stem cell to naivety
- PMID: 39134648
- PMCID: PMC11599042
- DOI: 10.1038/s41563-024-01971-4
Substrates mimicking the blastocyst geometry revert pluripotent stem cell to naivety
Abstract
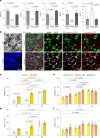
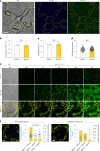
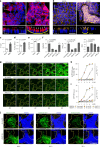
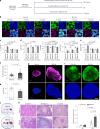
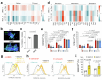
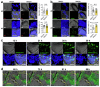
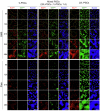
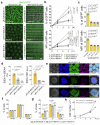
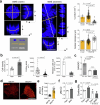
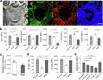
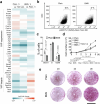
Naive pluripotent stem cells have the highest developmental potential but their in vivo existence in the blastocyst is transient. Here we report a blastocyst motif substrate for the in vitro reversion of mouse and human pluripotent stem cells to a naive state. The substrate features randomly varied microstructures, which we call motifs, mimicking the geometry of the blastocyst. Motifs representing mouse-blastocyst-scaled curvature ranging between 15 and 62 mm-1 were the most efficient in promoting reversion to naivety, as determined by time-resolved correlative analysis. In these substrates, apical constriction enhances E-cadherin/RAC1 signalling and activates the mechanosensitive nuclear transducer YAP, promoting the histone modification of pluripotency genes. This results in enhanced levels of pluripotency transcription factor NANOG, which persist even after cells are removed from the substrate. Pluripotent stem cells cultured in blastocyst motif substrates display a higher development potential in generating embryoid bodies and teratomas. These findings shed light on naivety-promoting substrate design and their large-scale implementation.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: A.L. and K.K. are inventors of a patent (WO 2011/054601 (A1)), related to cell culture inserts. X.X., Weiwei Wang and N.M. are inventors on a provisional patent application (EP 24160389.3 and 24161228.2) covering the biophysical methods for naivety reversion. The remaining authors declare no competing interests.
Figures





References
-
- Nichols, J. & Smith, A. I. Naive and primed pluripotent states. Cell Stem Cell4, 487–492 (2009). - PubMed
-
- Davidson, K. C., Mason, E. A. & Pera, M. F. The pluripotent state in mouse and human. Development142, 3090–3099 (2015). - PubMed
-
- Weinberger, L., Ayyash, M., Novershtern, N. & Hanna, J. H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol.17, 155–169 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- Program-oriented funding/Helmholtz Association
- I2B Funds/Helmholtz Association
- AIOrganoid/Helmholtz Association
- Helmholtz Cross Programme Initiative/Helmholtz Association
- VH-GS-503/Helmholtz Association
- 13GW0098/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- 13GW0099/Bundesministerium für Bildung und Forschung (Federal Ministry of Education and Research)
- No. 202206170040/China Scholarship Council (CSC)
- VH-VI-423/Helmholtz Virtuelles Institut Multifunktionale Biomaterialien für die Medizin (Helmholtz Virtual Institute Multifunctional Biomaterials for Medicine)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

