Staphylococcus aureus adapts to exploit collagen-derived proline during chronic infection
- PMID: 39134708
- PMCID: PMC11445067
- DOI: 10.1038/s41564-024-01769-9
Staphylococcus aureus adapts to exploit collagen-derived proline during chronic infection
Abstract
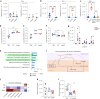
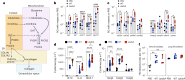
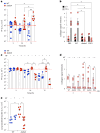
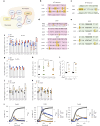
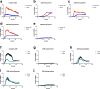
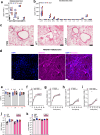
Staphylococcus aureus is a pulmonary pathogen associated with substantial human morbidity and mortality. As vaccines targeting virulence determinants have failed to be protective in humans, other factors are likely involved in pathogenesis. Here we analysed transcriptomic responses of human clinical isolates of S. aureus from initial and chronic infections. We observed upregulated collagenase and proline transporter gene expression in chronic infection isolates. Metabolomics of bronchiolar lavage fluid and fibroblast infection, growth assays and analysis of bacterial mutant strains showed that airway fibroblasts produce collagen during S. aureus infection. Host-adapted bacteria upregulate collagenase, which degrades collagen and releases proline. S. aureus then imports proline, which fuels oxidative metabolism via the tricarboxylic acid cycle. Proline metabolism provides host-adapted S. aureus with a metabolic benefit enabling out-competition of non-adapted strains. These data suggest that clinical settings characterized by airway repair processes and fibrosis provide a milieu that promotes S. aureus adaptation and supports infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Diep, B. A. & Otto, M. Methicillin-resistant Staphylococcus aureus infection is associated with increased mortality. Future Microbiol.7, 189–191 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

