JCVI: A versatile toolkit for comparative genomics analysis
- PMID: 39135687
- PMCID: PMC11316928
- DOI: 10.1002/imt2.211
JCVI: A versatile toolkit for comparative genomics analysis
Abstract
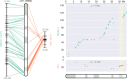
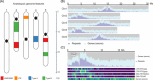
The life cycle of genome builds spans interlocking pillars of assembly, annotation, and comparative genomics to drive biological insights. While tools exist to address each pillar separately, there is a growing need for tools to integrate different pillars of a genome project holistically. For example, comparative approaches can provide quality control of assembly or annotation; genome assembly, in turn, can help to identify artifacts that may complicate the interpretation of genome comparisons. The JCVI library is a versatile Python-based library that offers a suite of tools that excel across these pillars. Featuring a modular design, the JCVI library provides high-level utilities for tasks such as format parsing, graphics generation, and manipulation of genome assemblies and annotations. Supporting genomics algorithms like MCscan and ALLMAPS are widely employed in building genome releases, producing publication-ready figures for quality assessment and evolutionary inference. Developed and maintained collaboratively, the JCVI library emphasizes quality and reusability.
Keywords: comparative genomics; genome annotation; genome assembly; genomic data; visualization.
© 2024 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
