Intestinal microbiota by angiotensin receptor blocker therapy exerts protective effects against hypertensive damages
- PMID: 39135690
- PMCID: PMC11316932
- DOI: 10.1002/imt2.222
Intestinal microbiota by angiotensin receptor blocker therapy exerts protective effects against hypertensive damages
Abstract
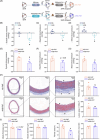
Dysbiosis of the gut microbiota has been implicated in hypertension, and drug-host-microbiome interactions have drawn considerable attention. However, the influence of angiotensin receptor blocker (ARB)-shaped gut microbiota on the host is not fully understood. In this work, we assessed the alterations of blood pressure (BP), vasculatures, and intestines following ARB-modified gut microbiome treatment and evaluated the changes in the intestinal transcriptome and serum metabolome in hypertensive rats. Hypertensive patients with well-controlled BP under ARB therapy were recruited as human donors, spontaneously hypertensive rats (SHRs) receiving normal saline or valsartan were considered animal donors, and SHRs were regarded as recipients. Histological and immunofluorescence staining was used to assess the aorta and small intestine, and 16S rRNA amplicon sequencing was performed to examine gut bacteria. Transcriptome and metabonomic analyses were conducted to determine the intestinal transcriptome and serum metabolome, respectively. Notably, ARB-modified fecal microbiota transplantation (FMT), results in marked decreases in systolic BP levels, collagen deposition and reactive oxygen species accumulation in the vasculature, and alleviated intestinal structure impairments in SHRs. These changes were linked with the reconstruction of the gut microbiota in SHR recipients post-FMT, especially with a decreased abundance of Lactobacillus, Aggregatibacter, and Desulfovibrio. Moreover, ARB-treated microbes contributed to increased intestinal Ciart, Per1, Per2, Per3, and Cipc gene levels and decreased Nfil3 and Arntl expression were detected in response to ARB-treated microbes. More importantly, circulating metabolites were dramatically reduced in ARB-FMT rats, including 6beta-Hydroxytestosterone and Thromboxane B2. In conclusion, ARB-modified gut microbiota exerts protective roles in vascular remodeling and injury, metabolic abnormality and intestinal dysfunctions, suggesting a pivotal role in mitigating hypertension and providing insights into the cross-talk between antihypertensive medicines and the gut microbiome.
Keywords: angiotensin receptor blockers; antihypertensive; gut microbiota; hypertension; vascular injury.
© 2024 The Author(s). iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Saladini, Francesca , Mancusi Costantino, Bertacchini Fabio, Spannella Francesco, Maloberti Alessandro, Giavarini Alessandra, and Rosticci Martina, et al. 2020. “Diagnosis and Treatment of Hypertensive Emergencies and Urgencies Among Italian Emergency and Intensive Care Departments. Results From an Italian Survey: Progetto GEAR (Gestione Dell'emergenza E Urgenza in ARea Critica).” European Journal of Internal Medicine 71: 50–56. 10.1016/j.ejim.2019.10.004 - DOI - PubMed
-
- Zhou, Bin , Bentham James, Di Cesare Mariachiara, Bixby Honor, Danaei Goodarz, Cowan Melanie J., and Paciorek Christopher J., et al. 2017. “Worldwide Trends in Blood Pressure from 1975 to 2015: A Pooled Analysis of 1479 Population‐Based Measurement Studies with 19·1 Million Participants.” The Lancet 389: 37–55. 10.1016/s0140-6736(16)31919-5 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
