Review of ABO Expression and Variations based on Transcriptional Regulation of the ABO Blood Group Gene
- PMID: 39135854
- PMCID: PMC11318969
- DOI: 10.1159/000536556
Review of ABO Expression and Variations based on Transcriptional Regulation of the ABO Blood Group Gene
Abstract
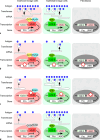
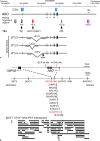
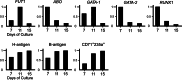
Background and summary: We review the transcriptional regulation of ABO expression and discuss variants in the promoter and erythroid cell-specific regulatory region in individuals with weak ABO phenotypes such as Bm, Am, B3, and A3. We also review the molecular mechanisms responsible for variations in ABO expression in development and disease including the cell type-specific expression of ABO during erythroid cell differentiation, and reduction of A- or B-antigens in cancer cells or on red blood cells in patients with leukemia. Although the relationship between ABO blood group antigens and diseases has been characterized, the physiological significance of the ABO blood group system remains unclear.
Key messages: This review discusses accumulated knowledge of the ABO gene regulation and potential reasons for conservation of ABO during evolution.
Keywords: ABO blood group; ABO gene; Transcriptional regulation; Weak ABO phenotype.
© 2024 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



Similar articles
-
Presence of nucleotide substitutions in transcriptional regulatory elements such as the erythroid cell-specific enhancer-like element and the ABO promoter in individuals with phenotypes A3 and B3, respectively.Vox Sang. 2014 Aug;107(2):171-80. doi: 10.1111/vox.12136. Epub 2014 Mar 6. Vox Sang. 2014. PMID: 24602004
-
Presence of nucleotide substitutions in the ABO promoter in individuals with phenotypes A3 and B3.Vox Sang. 2016 Apr;110(3):285-7. doi: 10.1111/vox.12363. Epub 2015 Nov 3. Vox Sang. 2016. PMID: 26529276
-
Blood group B gene is barely expressed in in vitro erythroid culture of Bm-derived CD34+ cells without an erythroid cell-specific regulatory element.Vox Sang. 2015 Apr;108(3):302-9. doi: 10.1111/vox.12220. Epub 2014 Dec 18. Vox Sang. 2015. PMID: 25523606
-
ABO blood-group antigens in oral cancer.J Dent Res. 2005 Jan;84(1):21-8. doi: 10.1177/154405910508400103. J Dent Res. 2005. PMID: 15615870 Review.
-
Regulation of ABO gene expression.Leg Med (Tokyo). 2005 Jul;7(4):263-5. doi: 10.1016/j.legalmed.2004.10.009. Leg Med (Tokyo). 2005. PMID: 15939654 Review.
Cited by
-
Regulation of Blood Group Expression: Another Layer of Complexity to Consider.Transfus Med Hemother. 2024 Jul 2;51(4):207-209. doi: 10.1159/000539611. eCollection 2024 Aug. Transfus Med Hemother. 2024. PMID: 39135853 Free PMC article. No abstract available.
References
-
- Landsteiner K. Zur Kenntnis der antifermentativen, lytischen und agglutinierenden Wirkungen des Blutserums und der Lymphe. Zentralblatt Bakteriologie. 1900;27:357–62.
-
- LandsteinerÜber K. Agglutinationserscheinungen normalen menschlichen Blutes. Wien Klin Woschenschr. 1901;14:1132–4.
-
- Daniels G. ABO, H, and Lewis systems. In: Daniels G, editor. Human blood groups. 3rd ed. West Sussex: Wiley-Blackwell; 2013. p. 11–95.
-
- Clausen H, White T, Takio K, Titani K, Stroud M, Holmes E, et al. . Isolation to homogeneity and partial characterization of a histo-blood group A defined Fucα1→2Galα1→3-N-acetylgalactosaminyltransferase from human lung tissue. J Biol Chem. 1990;265:1139‒45. - PubMed
-
- Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345(6272):229–33. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

