Species diversity in Pseudocercospora
- PMID: 39135885
- PMCID: PMC11317867
- DOI: 10.3114/fuse.2024.13.03
Species diversity in Pseudocercospora
Abstract
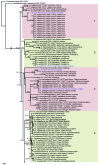
Species of Pseudocercospora are commonly associated with leaf and fruit spots on diverse plant hosts in sub-tropical and tropical regions. Pseudocercospora spp. have mycosphaerella-like sexual morphs, but represent a distinct genus in Mycosphaerellaceae (Mycosphaerellales, Dothideomycetes). The present study adds a further 29 novel species of Pseudocercospora from 413 host species representing 297 host genera occurring in 60 countries and designates four epitypes and one lectotype for established names. This study recognises 329 species names, with an additional 69 phylogenetic lineages remaining unnamed due to difficulty in being able to unambiguously apply existing names to those lineages. To help elucidate the taxonomy of these species, a phylogenetic tree was generated from multi-locus DNA sequence data of the internal transcribed spacers and intervening 5.8S nuclear nrRNA gene (ITS), partial actin (actA), and partial translation elongation factor 1-alpha (tef1), as well as the partial DNA-directed RNA polymerase II second largest subunit (rpb2) gene sequences. Novel species described in this study include those from various countries as follows: Australia, Ps. acaciicola from leaf spots on Acacia sp., Ps. anopter from leaf spots on Anopterus glandulosus, Ps. asplenii from leaf spots on Asplenium dimorphum, Ps. australiensis from leaf spots on Eucalyptus gunnii, Ps. badjensis from leaf spots on Eucalyptus badjensis, Ps. erythrophloeicola from leaf spots on Erythrophleum chlorostachys, Ps. grevilleae from leaf spots on Grevillea sp., Ps. lophostemonigena from leaf spots on Lophostemon confertus, Ps. lophostemonis from leaf spots on Lophostemon lactifluus, Ps. paramacadamiae from leaf spots on Macadamia integrifolia, Ps. persooniae from leaf spots on Persoonia sp., Ps. pultenaeae from leaf spots on Pultenaea daphnoides, Ps. tristaniopsidis from leaf spots on Tristaniopsis collina, Ps. victoriae from leaf spots on Eucalyptus globoidea. Brazil, Ps. musigena from leaf spots on Musa sp. China, Ps. lonicerae-japonicae from leaf spots on Lonicera japonica, Ps. rubigena leaf spots on Rubus sp. France (Réunion), Ps. wingfieldii from leaf spots on Acacia heterophylla. Malaysia, Ps. musarum from leaf spots on Musa sp. Netherlands, Ps. rhododendri from leaf spots on Rhododendron sp. South Africa, Ps. balanitis from leaf spots on Balanites sp., Ps. dovyalidicola from leaf spots on Dovyalis zeyheri, Ps. encephalarticola from leaf spots on Encephalartos sp. South Korea, Ps. grewiana from leaf spots on Grewia biloba, Ps. parakaki from leaf spots on Diospyros kaki, Ps. pseudocydoniae from leaf spots on Chaenomeles lagenaria, Ps. paracydoniae from leaf spots on Chaenomeles speciosa. Thailand, Ps. acerigena from leaf spots on Acer sp., Ps. tectonigena from leaf spots on Tectona grandis. Epitypes are designated for Cercospora bonjeaneae-rectae, Cercospora halleriae, Ps. eucleae, and an epitype as well as a lectotype for Ps. macadamiae. Results obtained in the present study contribute to a better understanding of the host specificity and distribution in Pseudocercospora spp., many of which represent important pathogens of food or fibre crops, or organisms of quarantine concern. Citation: Groenewald JZ, Chen YY, Zhang Y, Roux J, Shin H-D, Shivas RG, Summerell BA, Braun U, Alfenas AC, Ujat AH, Nakashima C, Crous PW (2024). Species diversity in Pseudocercospora. Fungal Systematics and Evolution 13: 29-89. doi: 10.3114/fuse.2024.13.03.
Keywords: Multi-gene phylogeny Mycosphaerellaceae new taxa; plant pathogen taxonomy.
© 2024 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

































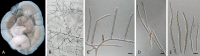
































References
-
- Acabal BD, Jr, Groenewald JZ, Crous PW, et al. (2014). First report of Pseudocercospora jahnii in the Philippines. Australasian Plant Disease Notes 9: 148.
-
- Akinsanmi OA, Carvalhais LC. (2020). Draft genome of the macadamia husk spot pathogen, Pseudocercospora macadamiae . Phytopathology 110: 1503–1506. - PubMed
-
- Bakhshi M, Arzanlou M, Babai-Ahari A.et al. (2014). Multi-gene analysis of Pseudocercospora spp. from Iran. Phytotaxa 184: 245–264.
-
- Beilharz V, Mayers PE, Pascoe IG. (2003). Pseudocercospora macadamiae sp. nov., the cause of husk spot of macadamia. Australasian Plant Pathology 32: 279–282.
-
- Braun U. (1995). A monograph of Cercosporella, Ramularia and allied genera (phytopathogenic hyphomycetes). Vol. 1. Eching: IHW Verlag.
LinkOut - more resources
Full Text Sources
Miscellaneous
