The longitudinal characterization of immune responses in COVID-19 patients reveals novel prognostic signatures for disease severity, patients' survival and long COVID
- PMID: 39136010
- PMCID: PMC11317765
- DOI: 10.3389/fimmu.2024.1381091
The longitudinal characterization of immune responses in COVID-19 patients reveals novel prognostic signatures for disease severity, patients' survival and long COVID
Abstract
Introduction: SARS-CoV-2 pandemic still poses a significant burden on global health and economy, especially for symptoms persisting beyond the acute disease. COVID-19 manifests with various degrees of severity and the identification of early biomarkers capable of stratifying patient based on risk of progression could allow tailored treatments.
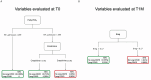
Methods: We longitudinally analyzed 67 patients, classified according to a WHO ordinal scale as having Mild, Moderate, or Severe COVID-19. Peripheral blood samples were prospectively collected at hospital admission and during a 6-month follow-up after discharge. Several subsets and markers of the innate and adaptive immunity were monitored as putative factors associated with COVID-19 symptoms.
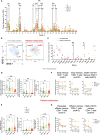
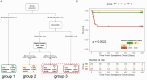
Results: More than 50 immunological parameters were associated with disease severity. A decision tree including the main clinical, laboratory, and biological variables at admission identified low NK-cell precursors and CD14+CD91+ monocytes, and high CD8+ Effector Memory T cell frequencies as the most robust immunological correlates of COVID-19 severity and reduced survival. Moreover, low regulatory B-cell frequency at one month was associated with the susceptibility to develop long COVID at six months, likely due to their immunomodulatory ability.
Discussion: These results highlight the profound perturbation of the immune response during COVID-19. The evaluation of specific innate and adaptive immune-cell subsets allows to distinguish between different acute and persistent COVID-19 symptoms.
Keywords: COVID-19 patients’ survival; COVID-19 severity; SARS-CoV-2 adaptive immunity; SARS-CoV-2 innate immunity; long COVID.
Copyright © 2024 Noviello, De Lorenzo, Chimienti, Maugeri, De Lalla, Siracusano, Lorè, Rancoita, Cugnata, Tassi, Dispinseri, Abbati, Beretta, Ruggiero, Manfredi, Merolla, Cantarelli, Tresoldi, Pastori, Caccia, Sironi, Marzinotto, Saliu, Ghezzi, Lampasona, Vicenzi, Cinque, Manfredi, Scarlatti, Dellabona, Lopalco, Di Serio, Malnati, Ciceri, Rovere-Querini and Bonini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
-
- Corradini P, Agrati C, Apolone G, Mantovani A, Giannarelli D, Marasco V, et al. . Humoral and T-cell immune response after 3 doses of messenger RNA severe acute respiratory syndrome coronavirus 2 vaccines in fragile patients: the italian VAX4FRAIL study. Clin Infect Dis. (2023) 76:426–38. doi: 10.1093/cid/ciac404 - DOI - PMC - PubMed
-
- Wouters OJ, Shadlen KC, Salcher-konrad M, Pollard AJ, Larson HJ, Teerawattananon Y, et al. . Health Policy Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet. (2021) 397:1023–34. doi: 10.1016/S0140-6736(21)00306-8 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

