Targeting Neuraminidase 4 Attenuates Kidney Fibrosis in Mice
- PMID: 39136142
- PMCID: PMC11497051
- DOI: 10.1002/advs.202406936
Targeting Neuraminidase 4 Attenuates Kidney Fibrosis in Mice
Abstract
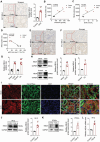
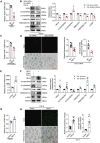
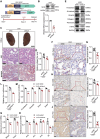
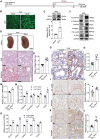
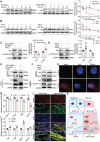
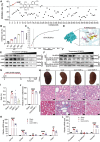
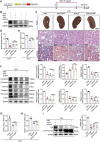
Despite significant progress in therapy, there remains a lack of substantial evidence regarding the molecular factors that lead to renal fibrosis. Neuraminidase 4 (NEU4), an enzyme that removes sialic acids from glycoconjugates, has an unclear role in chronic progressive fibrosis. Here, this study finds that NEU4 expression is markedly upregulated in mouse fibrotic kidneys induced by folic acid or unilateral ureter obstruction, and this elevation is observed in patients with renal fibrosis. NEU4 knockdown specifically in the kidney attenuates the epithelial-to-mesenchymal transition, reduces the production of pro-fibrotic cytokines, and decreases cellular senescence in male mice. Conversely, NEU4 overexpression exacerbates the progression of renal fibrosis. Mechanistically, NEU4254-388aa interacts with Yes-associated protein (YAP) at WW2 domain (231-263aa), promoting its nucleus translocation and activation of target genes, thereby contributing to renal fibrosis. 3,5,6,7,8,3',4'-Heptamethoxyflavone, a natural compound, is identified as a novel NEU4 inhibitor, effectively protecting mice from renal fibrosis in a NEU4-dependent manner. Collectively, the findings suggest that NEU4 may represent a promising therapeutic target for kidney fibrosis.
Keywords: 3,5,6,7,8,3ʹ,4ʹ‐Heptamethoxyflavone; NEU4; YAP; renal fibrosis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
