CDK4/6 Inhibitors Impede Chemoresistance and Inhibit Tumor Growth of Small Cell Lung Cancer
- PMID: 39136283
- PMCID: PMC11481398
- DOI: 10.1002/advs.202400666
CDK4/6 Inhibitors Impede Chemoresistance and Inhibit Tumor Growth of Small Cell Lung Cancer
Abstract
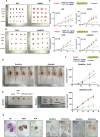
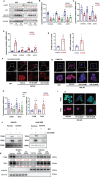
Small cell lung cancer (SCLC) is characterized by rapid development of chemoresistance and poor outcomes. Cyclin-dependent kinase 4/6 inhibitors (CDK4/6is) are widely used in breast cancer and other cancer types. However, the molecular mechanisms of CDK4/6 in SCLC chemoresistance remain poorly understood. Here, Rb1flox/flox, Trp53flox/flox, Ptenflox/flox (RTP) and Rb1flox/flox, Trp53flox/flox, MycLSL/LSL (RPM) spontaneous SCLC mouse models, SCLC cell line-derived xenograft (CDX) models, and SCLC patient-derived xenograft (PDX) models are established to reveal the potential effects of CDK4/6is on SCLC chemoresistance. In this study, it is found that CDK4/6is palbociclib (PD) or ribociclib (LEE) combined with chemotherapeutic drugs significantly inhibit SCLC tumor growth. Mechanistically, CDK4/6is do not function through the classic Retionblastoma1 (RB) dependent axis in SCLC. CDK4/6is induce impair autophagy through the AMBRA1-lysosome signaling pathway. The upregulated AMBRA1 protein expression leads to CDK6 degradation via autophagy, and the following TFEB and TFE3 nuclear translocation inhibition leading to the lysosome-related genes levels downregulation. Moreover, it is found that the expression of CDK6 is higher in SCLC tumors than in normal tissue and it is associated with the survival and prognosis of SCLC patients. Finally, these findings demonstrate that combining CDK4/6is with chemotherapy treatment may serve as a potential therapeutic option for SCLC patients.
Keywords: AMBRA1; CDK6; SCLC; autophagic flux; chemoresistance; lysosomal depletion.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- a) Sabari J. K., Lok B. H., Laird J. H., Poirier J. T., Rudin C. M., Nat. Rev. Clin. Oncol. 2017, 14, 549; - PMC - PubMed
- b) Tsoukalas N., Aravantinou‐Fatorou E., Baxevanos P., Tolia M., Tsapakidis K., Galanopoulos M., Liontos M., Kyrgias G., Ann. Transl. Med. 2018, 6, 145; - PMC - PubMed
- c) van Meerbeeck J. P., Fennell D. A., De Ruysscher D. K., Lancet 2011, 378, 1741. - PubMed
-
- Kahnert K., Kauffmann‐Guerrero D., Huber R. M., Clin. Lung Cancer 2016, 17, 325. - PubMed
-
- a) Gazdar A. F., Bunn P. A., Minna J. D., Nat. Rev. Cancer 2017, 17, 725; - PubMed
- b) Belani C. P., Dahlberg S. E., Rudin C. M., Fleisher M., Chen H. X., Takebe N., Velasco M. R., Tester W. J., Sturtz K., Hann C. L., Shanks J. C., Monga M., Ramalingam S. S., Schiller J. H., Cancer 2016, 122, 2371. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 81972809/National Natural Science Foundation of China
- 82172769/National Natural Science Foundation of China
- 82102943/National Natural Science Foundation of China
- 81802254/National Natural Science Foundation of China
- 81902311/National Natural Science Foundation of China
- LC2016ZD029/Clinical Research Initiative Project of Southern Medical University
- 2019A030317022/Science and Technology Planning Project of Guangdong Province, China
- 2024A04J4876/Science and Technology Program of Guangzhou
- 202002030359/Guangzhou Science and Technology Project
- 2020A1515011454/Guangdong Natural Science Foundation
- 2021GGB003/Young and Middle-aged Talents Training Project of Fujian Provincial Health Commission
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
