Functional and practical insights into three lactococcal antiphage systems
- PMID: 39136492
- PMCID: PMC11409693
- DOI: 10.1128/aem.01120-24
Functional and practical insights into three lactococcal antiphage systems
Abstract
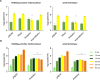
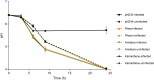
The persistent challenge of phages in dairy fermentations requires the development of starter cultures with enhanced phage resistance. Recently, three plasmid-encoded lactococcal antiphage systems, named Rhea, Aristaios, and Kamadhenu, were discovered. These systems were found to confer high levels of resistance against various Skunavirus members. In the present study, their effectiveness against phage infection was confirmed in milk-based medium, thus validating their potential to ensure reliable dairy fermentations. We furthermore demonstrated that Rhea and Kamadhenu do not directly hinder phage genome replication, transcription, or associated translation. Conversely, Aristaios was found to interfere with phage transcription. Two of the antiphage systems are encoded on pMRC01-like conjugative plasmids, and the Kamadhenu-encoding plasmid was successfully transferred by conjugation to three lactococcal strains, each of which acquired substantially enhanced phage resistance against Skunavirus members. Such advances in our knowledge of the lactococcal phage resistome and the possibility of mobilizing these protective functions to bolster phage protection in sensitive strains provide practical solutions to the ongoing phage problem in industrial food fermentations.IMPORTANCEIn the current study, we characterized and evaluated the mechanistic diversity of three recently described, plasmid-encoded lactococcal antiphage systems. These systems were found to confer high resistance against many members of the most prevalent and problematic lactococcal phage genus, rendering them of particular interest to the dairy industry, where persistent phage challenge requires the development of starter cultures with enhanced phage resistance characteristics. Our acquired knowledge highlights that enhanced understanding of lactococcal phage resistance systems and their encoding plasmids can provide rational and effective solutions to the enduring issue of phage infections in dairy fermentation facilities.
Keywords: Abi; Lactococcus; bacterial immunity; lactic acid bacteria; phage defense.
Conflict of interest statement
P.P.D.W., I.M.H.V.R., and N.N.M.E.V.P. work for dsm-firmenich.
Figures

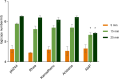


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

