Phase separation of PML/RARα and BRD4 coassembled microspeckles governs transcriptional dysregulation in acute promyelocytic leukemia
- PMID: 39136995
- PMCID: PMC11348160
- DOI: 10.1073/pnas.2406519121
Phase separation of PML/RARα and BRD4 coassembled microspeckles governs transcriptional dysregulation in acute promyelocytic leukemia
Abstract
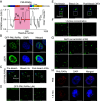
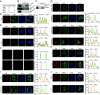
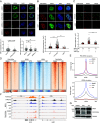
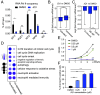
In acute promyelocytic leukemia (APL), the promyelocytic leukemia-retinoic acid receptor alpha (PML/RARα) fusion protein destroys PML nuclear bodies (NBs), leading to the formation of microspeckles. However, our understanding, largely learned from morphological observations, lacks insight into the mechanisms behind PML/RARα-mediated microspeckle formation and its role in APL leukemogenesis. This study presents evidence uncovering liquid-liquid phase separation (LLPS) as a key mechanism in the formation of PML/RARα-mediated microspeckles. This process is facilitated by the intrinsically disordered region containing a large portion of PML and a smaller segment of RARα. We demonstrate the coassembly of bromodomain-containing protein 4 (BRD4) within PML/RARα-mediated condensates, differing from wild-type PML-formed NBs. In the absence of PML/RARα, PML NBs and BRD4 puncta exist as two independent phases, but the presence of PML/RARα disrupts PML NBs and redistributes PML and BRD4 into a distinct phase, forming PML/RARα-assembled microspeckles. Genome-wide profiling reveals a PML/RARα-induced BRD4 redistribution across the genome, with preferential binding to super-enhancers and broad-promoters (SEBPs). Mechanistically, BRD4 is recruited by PML/RARα into nuclear condensates, facilitating BRD4 chromatin binding to exert transcriptional activation essential for APL survival. Perturbing LLPS through chemical inhibition (1, 6-hexanediol) significantly reduces chromatin co-occupancy of PML/RARα and BRD4, attenuating their target gene activation. Finally, a series of experimental validations in primary APL patient samples confirm that PML/RARα forms microspeckles through condensates, recruits BRD4 to coassemble condensates, and co-occupies SEBP regions. Our findings elucidate the biophysical, pathological, and transcriptional dynamics of PML/RARα-assembled microspeckles, underscoring the importance of BRD4 in mediating transcriptional activation that enables PML/RARα to initiate APL.
Keywords: BRD4; PML/RARα; acute promyelocytic leukemia; liquid–liquid phase separation; transcriptional dysregulation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Similar articles
-
A PML/RARα direct target atlas redefines transcriptional deregulation in acute promyelocytic leukemia.Blood. 2021 Mar 18;137(11):1503-1516. doi: 10.1182/blood.2020005698. Blood. 2021. PMID: 32854112 Free PMC article.
-
BRD4 interacts with PML/RARα in acute promyelocytic leukemia.Front Med. 2018 Dec;12(6):726-734. doi: 10.1007/s11684-017-0604-x. Epub 2018 Dec 14. Front Med. 2018. PMID: 30552662
-
Caspases mediate retinoic acid-induced degradation of the acute promyelocytic leukemia PML/RARalpha fusion protein.Blood. 1998 Oct 1;92(7):2244-51. Blood. 1998. PMID: 9746761
-
The functional roles of PML nuclear bodies in genome maintenance.Mutat Res. 2018 May;809:99-107. doi: 10.1016/j.mrfmmm.2017.05.002. Epub 2017 May 5. Mutat Res. 2018. PMID: 28521962 Review.
-
Transcriptional regulation in acute promyelocytic leukemia.Oncogene. 2001 Oct 29;20(49):7204-15. doi: 10.1038/sj.onc.1204853. Oncogene. 2001. PMID: 11704848 Review.
Cited by
-
Biomolecular condensates in immune cell fate.Nat Rev Immunol. 2025 Jun;25(6):445-459. doi: 10.1038/s41577-025-01130-z. Epub 2025 Jan 28. Nat Rev Immunol. 2025. PMID: 39875604 Review.
-
Liquid-liquid phase separation in normal hematopoiesis and hematological diseases.Cell Tissue Res. 2025 Jul;401(1):15-27. doi: 10.1007/s00441-025-03974-2. Epub 2025 May 17. Cell Tissue Res. 2025. PMID: 40381034 Review.
References
-
- Dyck J. A., et al. , A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell 76, 333–343 (1994). - PubMed
-
- Weis K., et al. , Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell 76, 345–356 (1994). - PubMed
MeSH terms
Substances
Grants and funding
- 32170663/National Natural Science Foundation of China
- 82350710226/National Natural Science Foundation of China
- 82370178/National Natural Science Foundation of China
- 2023YFA1800401/National Key R&D Program of China
- dy2yhbrc202002/Young Reserve Talent Program for the Second Hospital of Dalian Medical University
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

