Pediatric glioma immune profiling identifies TIM3 as a therapeutic target in BRAF fusion pilocytic astrocytoma
- PMID: 39137048
- PMCID: PMC11444160
- DOI: 10.1172/JCI177413
Pediatric glioma immune profiling identifies TIM3 as a therapeutic target in BRAF fusion pilocytic astrocytoma
Abstract
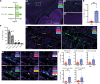
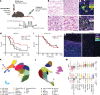
Despite being the leading cause of cancer-related childhood mortality, pediatric gliomas have been relatively understudied, and the repurposing of immunotherapies has not been successful. Whole-transcriptome sequencing, single-cell sequencing, and sequential multiplex immunofluorescence were used to identify an immunotherapeutic strategy that could be applied to multiple preclinical glioma models. MAPK-driven pediatric gliomas have a higher IFN signature relative to other molecular subgroups. Single-cell sequencing identified an activated and cytotoxic microglia (MG) population designated MG-Act in BRAF-fused, MAPK-activated pilocytic astrocytoma (PA), but not in high-grade gliomas or normal brain. T cell immunoglobulin and mucin domain 3 (TIM3) was expressed on MG-Act and on the myeloid cells lining the tumor vasculature but not normal brain vasculature. TIM3 expression became upregulated on immune cells in the PA microenvironment, and anti-TIM3 reprogrammed ex vivo immune cells from human PAs to a proinflammatory cytotoxic phenotype. In a genetically engineered murine model of MAPK-driven, low-grade gliomas, anti-TIM3 treatment increased median survival over IgG- and anti-PD-1-treated mice. Single-cell RNA-Seq data during the therapeutic window of anti-TIM3 revealed enrichment of the MG-Act population. The therapeutic activity of anti-TIM3 was abrogated in mice on the CX3CR1 MG-KO background. These data support the use of anti-TIM3 in clinical trials of pediatric low-grade, MAPK-driven gliomas.
Keywords: Brain cancer; Cancer immunotherapy; Immunology; Oncology.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

