Arl2 GTPase associates with the centrosomal protein Cdk5rap2 to regulate cortical development via microtubule organization
- PMID: 39137170
- PMCID: PMC11321591
- DOI: 10.1371/journal.pbio.3002751
Arl2 GTPase associates with the centrosomal protein Cdk5rap2 to regulate cortical development via microtubule organization
Abstract
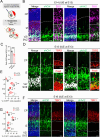
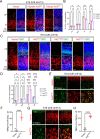
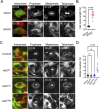
ADP ribosylation factor-like GTPase 2 (Arl2) is crucial for controlling mitochondrial fusion and microtubule assembly in various organisms. Arl2 regulates the asymmetric division of neural stem cells in Drosophila via microtubule growth. However, the function of mammalian Arl2 during cortical development was unknown. Here, we demonstrate that mouse Arl2 plays a new role in corticogenesis via regulating microtubule growth, but not mitochondria functions. Arl2 knockdown (KD) leads to impaired proliferation of neural progenitor cells (NPCs) and neuronal migration. Arl2 KD in mouse NPCs significantly diminishes centrosomal microtubule growth and delocalization of centrosomal proteins Cdk5rap2 and γ-tubulin. Moreover, Arl2 physically associates with Cdk5rap2 by in silico prediction using AlphaFold multimer, which was validated by co-immunoprecipitation and proximity ligation assay. Remarkably, Cdk5rap2 overexpression significantly rescues the neurogenesis defects caused by Arl2 KD. Therefore, Arl2 plays an important role in mouse cortical development through microtubule growth via the centrosomal protein Cdk5rap2.
Copyright: © 2024 Ma et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

