Candida albicans' inorganic phosphate transport and evolutionary adaptation to phosphate scarcity
- PMID: 39137212
- PMCID: PMC11343460
- DOI: 10.1371/journal.pgen.1011156
Candida albicans' inorganic phosphate transport and evolutionary adaptation to phosphate scarcity
Abstract
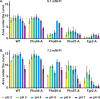
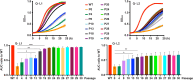
Phosphorus is essential in all cells' structural, metabolic and regulatory functions. For fungal cells that import inorganic phosphate (Pi) up a steep concentration gradient, surface Pi transporters are critical capacitators of growth. Fungi must deploy Pi transporters that enable optimal Pi uptake in pH and Pi concentration ranges prevalent in their environments. Single, triple and quadruple mutants were used to characterize the four Pi transporters we identified for the human fungal pathogen Candida albicans, which must adapt to alkaline conditions during invasion of the host bloodstream and deep organs. A high-affinity Pi transporter, Pho84, was most efficient across the widest pH range while another, Pho89, showed high-affinity characteristics only within one pH unit of neutral. Two low-affinity Pi transporters, Pho87 and Fgr2, were active only in acidic conditions. Only Pho84 among the Pi transporters was clearly required in previously identified Pi-related functions including Target of Rapamycin Complex 1 signaling, oxidative stress resistance and hyphal growth. We used in vitro evolution and whole genome sequencing as an unbiased forward genetic approach to probe adaptation to prolonged Pi scarcity of two quadruple mutant lineages lacking all 4 Pi transporters. Lineage-specific genomic changes corresponded to divergent success of the two lineages in fitness recovery during Pi limitation. Initial, large-scale genomic alterations like aneuploidies and loss of heterozygosity eventually resolved, as populations gained small-scale mutations. Severity of some phenotypes linked to Pi starvation, like cell wall stress hypersensitivity, decreased in parallel to evolving populations' fitness recovery in Pi scarcity, while severity of others like membrane stress responses diverged from Pi scarcity fitness. Among preliminary candidate genes for contributors to fitness recovery, those with links to TORC1 were overrepresented. Since Pi homeostasis differs substantially between fungi and humans, adaptive processes to Pi deprivation may harbor small-molecule targets that impact fungal growth, stress resistance and virulence.
Copyright: © 2024 Acosta-Zaldívar et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Update of
-
Candida albicans' inorganic phosphate transport and evolutionary adaptation to phosphate scarcity.bioRxiv [Preprint]. 2024 Feb 1:2024.01.29.577887. doi: 10.1101/2024.01.29.577887. bioRxiv. 2024. Update in: PLoS Genet. 2024 Aug 13;20(8):e1011156. doi: 10.1371/journal.pgen.1011156. PMID: 38352318 Free PMC article. Updated. Preprint.
References
-
- Garg A, Sanchez AM, Miele M, Schwer B, Shuman S. Cellular responses to long-term phosphate starvation of fission yeast: Maf1 determines fate choice between quiescence and death associated with aberrant tRNA biogenesis. Nucleic acids research. 2023;51(7):3094–115. Epub 2023/02/17. doi: 10.1093/nar/gkad063 ; PubMed Central PMCID: PMC10123115. - DOI - PMC - PubMed
-
- Ducousso-Detrez A, Fontaine J, Lounes-Hadj Sahraoui A, Hijri M. Diversity of Phosphate Chemical Forms in Soils and Their Contributions on Soil Microbial Community Structure Changes. Microorganisms. 2022;10(3). Epub 2022/03/27. doi: 10.3390/microorganisms10030609 ; PubMed Central PMCID: PMC8950675. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

