The dynamics and longevity of circulating CD4+ memory T cells depend on cell age and not the chronological age of the host
- PMID: 39137219
- PMCID: PMC11321570
- DOI: 10.1371/journal.pbio.3002380
The dynamics and longevity of circulating CD4+ memory T cells depend on cell age and not the chronological age of the host
Abstract
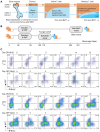
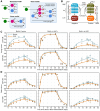
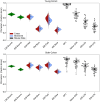
Quantifying the kinetics with which memory T cell populations are generated and maintained is essential for identifying the determinants of the duration of immunity. The quality and persistence of circulating CD4 effector memory (TEM) and central memory (TCM) T cells in mice appear to shift with age, but it is unclear whether these changes are driven by the aging host environment, by cell age effects, or both. Here, we address these issues by combining DNA labelling methods, established fate-mapping systems, a novel reporter mouse strain, and mathematical models. Together, these allow us to quantify the dynamics of both young and established circulating memory CD4 T cell subsets, within both young and old mice. We show that that these cells and their descendents become more persistent the longer they reside within the TCM and TEM pools. This behaviour may limit memory CD4 T cell diversity by skewing TCR repertoires towards clones generated early in life, but may also compensate for functional defects in new memory cells generated in old age.
Copyright: © 2024 Bullock et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
The dynamics and longevity of circulating CD4+ memory T cells depend on cell age and not the chronological age of the host.bioRxiv [Preprint]. 2024 Jun 25:2023.10.16.562650. doi: 10.1101/2023.10.16.562650. bioRxiv. 2024. Update in: PLoS Biol. 2024 Aug 13;22(8):e3002380. doi: 10.1371/journal.pbio.3002380. PMID: 38948729 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

