Testing the PEST hypothesis using relevant Rett mutations in MeCP2 E1 and E2 isoforms
- PMID: 39137370
- PMCID: PMC11540922
- DOI: 10.1093/hmg/ddae119
Testing the PEST hypothesis using relevant Rett mutations in MeCP2 E1 and E2 isoforms
Erratum in
-
Correction to: Testing the PEST hypothesis using relevant Rett mutations in MeCP2 E1 and E2 isoforms.Hum Mol Genet. 2025 Jun 4;34(12):1088. doi: 10.1093/hmg/ddaf072. Hum Mol Genet. 2025. PMID: 40323278 Free PMC article. No abstract available.
Abstract
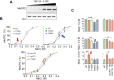
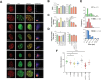
Mutations in methyl-CpG binding protein 2 (MeCP2), such as the T158M, P152R, R294X, and R306C mutations, are responsible for most Rett syndrome (RTT) cases. These mutations often result in altered protein expression that appears to correlate with changes in the nuclear size; however, the molecular details of these observations are poorly understood. Using a C2C12 cellular system expressing human MeCP2-E1 isoform as well as mouse models expressing these mutations, we show that T158M and P152R result in a decrease in MeCP2 protein, whereas R306C has a milder variation, and R294X resulted in an overall 2.5 to 3 fold increase. We also explored the potential involvement of the MeCP2 PEST domains in the proteasome-mediated regulation of MeCP2. Finally, we used the R294X mutant to gain further insight into the controversial competition between MeCP2 and histone H1 in the chromatin context. Interestingly, in R294X, MeCP2 E1 and E2 isoforms were differently affected, where the E1 isoform contributes to much of the overall protein increase observed, while E2 decreases by half. The modes of MeCP2 regulation, thus, appear to be differently regulated in the two isoforms.
Keywords: MeCP2; PEST sequences; Rett; chromatin; methyl CpG binding protein.
© The Author(s) 2024. Published by Oxford University Press.
Figures




References
-
- Lewis JD, Meehan RR, Henzel WJ. et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992;69:905–914. - PubMed
-
- Mnatzakanian GN, Lohi H, Munteanu I. et al. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet 2004;36:339–341. - PubMed
-
- Nan X, Ng HH, Johnson CA. et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998;393:386–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

