Interleukin-21 engineering enhances NK cell activity against glioblastoma via CEBPD
- PMID: 39137729
- PMCID: PMC11370652
- DOI: 10.1016/j.ccell.2024.07.007
Interleukin-21 engineering enhances NK cell activity against glioblastoma via CEBPD
Abstract
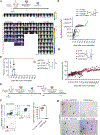
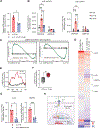
Glioblastoma (GBM) is an aggressive brain cancer with limited therapeutic options. Natural killer (NK) cells are innate immune cells with strong anti-tumor activity and may offer a promising treatment strategy for GBM. We compared the anti-GBM activity of NK cells engineered to express interleukin (IL)-15 or IL-21. Using multiple in vivo models, IL-21 NK cells were superior to IL-15 NK cells both in terms of safety and long-term anti-tumor activity, with locoregionally administered IL-15 NK cells proving toxic and ineffective at tumor control. IL-21 NK cells displayed a unique chromatin accessibility signature, with CCAAT/enhancer-binding proteins (C/EBP), especially CEBPD, serving as key transcription factors regulating their enhanced function. Deletion of CEBPD resulted in loss of IL-21 NK cell potency while its overexpression increased NK cell long-term cytotoxicity and metabolic fitness. These results suggest that IL-21, through C/EBP transcription factors, drives epigenetic reprogramming of NK cells, enhancing their anti-tumor efficacy against GBM.
Keywords: CEBPD; GBM; IL-21; NK cells.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.S., M. Daher, R.B., H.R., S.A., Y.L., N.U., E.L., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical. R.B., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Affimed GmbH. K.R. participates on the Scientific Advisory Board for Avenge Bio, Virogin Biotech, Navan Technologies, Caribou Biosciences, Bit Bio Limited, Replay Holdings, oNKo Innate, and The Alliance for Cancer Gene Therapy ACGT. K.R. is the Scientific founder of Syena. E.J.S. has served on the Scientific Advisory Board for Adaptimmune, Axio, Celaid, FibroBiologics, Navan Technologies, New York Blood Center and Novartis. M Daher participates on the Scientific Advisory Board for Cellsbin. M.S., H.S., E.J.S., and K.R. have filed for a patent (20230074303); “Cell immunotherapy for the treatment of cancer.”
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

