RIPK3 and caspase-8 interpret cytokine signals to regulate ILC3 survival in the gut
- PMID: 39137882
- PMCID: PMC11637958
- DOI: 10.1016/j.mucimm.2024.08.004
RIPK3 and caspase-8 interpret cytokine signals to regulate ILC3 survival in the gut
Erratum in
-
Corrigendum "RIPK3 and Caspase-8 interpret cytokine signals to regulate ILC3 survival in the gut". [Mucosal Immunol. 17/6 (2024) 1212-1221].Mucosal Immunol. 2025 Dec;18(6):1464-1465. doi: 10.1016/j.mucimm.2025.11.002. Epub 2025 Nov 14. Mucosal Immunol. 2025. PMID: 41241599 No abstract available.
Abstract
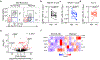
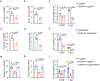
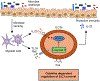
Group 3 innate lymphoid cells (ILC3s) are abundant in the developing or healthy intestine to critically support tissue homeostasis in response to microbial colonization. However, intestinal ILC3s are reduced during chronic infections, colorectal cancer, or inflammatory bowel disease (IBD), and the mechanisms driving these alterations remain poorly understood. Here we employed RNA sequencing of ILC3s from IBD patients and observed a significant upregulation of RIPK3, the central regulator of necroptosis, during intestinal inflammation. This was modeled in mice where we found that intestinal ILC3s express RIPK3, with conventional (c)ILC3s exhibiting high RIPK3 and low levels of pro-survival genes relative to lymphoid tissue inducer (LTi)-like ILC3s. ILC3-specific RIPK3 is promoted by gut microbiota, further upregulated following enteric infection, and dependent upon IL-23R and STAT3 signaling. However, lineage-specific deletion of RIPK3 revealed a redundant role in ILC3 survival, due to a blockade of RIPK3-mediated necroptosis by caspase 8, which was also activated in response to enteric infection. In contrast, lineage-specific deletion of caspase 8 resulted in loss of cILC3s from the healthy intestine and all ILC3 subsets during enteric infection, which increased pathogen burdens and gut inflammation. This function of caspase 8 required catalytic activity induced by TNF or TL1A and was dispensable if RIPK3 was simultaneously deleted. Caspase 8 activation and cell death were associated with increased Fas on ILC3s, and the Fas-FasL pathway was upregulated by cILC3s during enteric infection, which could restrain the abundance of intestinal ILC3s. Collectively, these data reveal that interpretation of key cytokine signals controls ILC3 survival following microbial challenge, and that an imbalance of these pathways, such as in IBD or across ILC3 subsets, provokes depletion of tissue-protective ILC3s from the inflamed intestine.
Keywords: Cytokines; IBD; Innate immunity; Innate lymphoid cells; Mucosal inflammation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: G.F.S. is on the Editorial Board of Mucosal Immunology. No other authors have a conflict of interest.
Figures



References
-
- Artis D, Spits H. The biology of innate lymphoid cells. Nature 2015; 517(7534): 293–301. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

