GRK2 kinases in the primary cilium initiate SMOOTHENED-PKA signaling in the Hedgehog cascade
- PMID: 39138140
- PMCID: PMC11322411
- DOI: 10.1371/journal.pbio.3002685
GRK2 kinases in the primary cilium initiate SMOOTHENED-PKA signaling in the Hedgehog cascade
Abstract
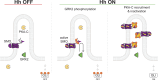
During Hedgehog (Hh) signal transduction in development and disease, the atypical G protein-coupled receptor (GPCR) SMOOTHENED (SMO) communicates with GLI transcription factors by binding the protein kinase A catalytic subunit (PKA-C) and physically blocking its enzymatic activity. Here, we show that GPCR kinase 2 (GRK2) orchestrates this process during endogenous mouse and zebrafish Hh pathway activation in the primary cilium. Upon SMO activation, GRK2 rapidly relocalizes from the ciliary base to the shaft, triggering SMO phosphorylation and PKA-C interaction. Reconstitution studies reveal that GRK2 phosphorylation enables active SMO to bind PKA-C directly. Lastly, the SMO-GRK2-PKA pathway underlies Hh signal transduction in a range of cellular and in vivo models. Thus, GRK2 phosphorylation of ciliary SMO and the ensuing PKA-C binding and inactivation are critical initiating events for the intracellular steps in Hh signaling. More broadly, our study suggests an expanded role for GRKs in enabling direct GPCR interactions with diverse intracellular effectors.
Copyright: © 2024 Walker et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: S.S. is the founder and scientific advisor of 7TM Antibodies GmbH, Jena, Germany. F.N. is an employee of 7TM Antibodies. All other authors declare no competing interests.
Figures



Update of
-
GRK2 Kinases in the Primary Cilium Initiate SMOOTHENED-PKA Signaling in the Hedgehog Cascade.bioRxiv [Preprint]. 2024 Apr 18:2023.05.10.540226. doi: 10.1101/2023.05.10.540226. bioRxiv. 2024. Update in: PLoS Biol. 2024 Aug 13;22(8):e3002685. doi: 10.1371/journal.pbio.3002685. PMID: 37214942 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

