Insights into the Interaction Landscape of the EVH1 Domain of Mena
- PMID: 39138154
- PMCID: PMC12395540
- DOI: 10.1021/acs.biochem.4c00331
Insights into the Interaction Landscape of the EVH1 Domain of Mena
Abstract
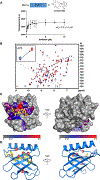
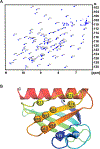
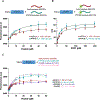
The Enabled/VASP homology 1 (EVH1) domain is a small module that interacts with proline-rich stretches in its ligands and is found in various signaling and scaffolding proteins. Mena, the mammalian homologue of Ena, is involved in diverse actin-associated events, such as membrane dynamics, bacterial motility, and tumor intravasation and extravasation. Two-dimensional (2D) 1H-15N HSQC NMR was used to study Mena EVH1 binding properties, defining the amino acids involved in ligand recognition for the physiological ligands ActA and PCARE, and a synthetic polyproline-inspired small molecule (hereafter inhibitor 6c). Chemical shift perturbations indicated that proline-rich segments bind in the conserved EVH1 hydrophobic cleft. The PCARE-derived peptide elicited more perturbations compared to the ActA-derived peptide, consistent with a previous report of a structural alteration in the solvent-exposed β7-β8 loop. Unexpectedly, EVH1 and the proline-rich segment of PTP1B did not exhibit NMR chemical shift perturbations; however, the high-resolution crystal structure implicated the conserved EVH1 hydrophobic cleft in ligand recognition. Intrinsic steady-state fluorescence and fluorescence polarization assays indicate that residues outside the proline-rich segment enhance the ligand affinity for EVH1 (Kd = 3-8 μM). Inhibitor 6c displayed tighter binding (Kd ∼ 0.3 μM) and occupies the same EVH1 cleft as physiological ligands. These studies revealed that the EVH1 domain enhances ligand affinity through recognition of residues flanking the proline-rich segments. Additionally, a synthetic inhibitor binds more tightly to the EVH1 domain than natural ligands, occupying the same hydrophobic cleft.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Volkman BF; Prehoda KE; Scott JA; Peterson FC; Lim WA Structure of the N-WASP EVH1 Domain-WIP Complex. Cell 2002, 111 (4), 565–576. - PubMed
-
- Barzik M; Carl UD; Schubert WD; Frank R; Wehland J; Heinz DW The N-terminal domain of Homer/Vesl is a new class II EVH1 domain. J. Mol. Biol. 2001, 309 (1), 155–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

