TFEB activation hallmarks antigenic experience of B lymphocytes and directs germinal center fate decisions
- PMID: 39138218
- PMCID: PMC11322606
- DOI: 10.1038/s41467-024-51166-3
TFEB activation hallmarks antigenic experience of B lymphocytes and directs germinal center fate decisions
Abstract
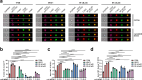
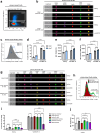
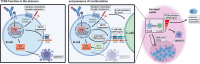
Ligation of the B cell antigen receptor (BCR) initiates humoral immunity. However, BCR signaling without appropriate co-stimulation commits B cells to death rather than to differentiation into immune effector cells. How BCR activation depletes potentially autoreactive B cells while simultaneously primes for receiving rescue and differentiation signals from cognate T lymphocytes remains unknown. Here, we use a mass spectrometry-based proteomic approach to identify cytosolic/nuclear shuttling elements and uncover transcription factor EB (TFEB) as a central BCR-controlled rheostat that drives activation-induced apoptosis, and concurrently promotes the reception of co-stimulatory rescue signals by supporting B cell migration and antigen presentation. CD40 co-stimulation prevents TFEB-driven cell death, while enhancing and prolonging TFEB's nuclear residency, which hallmarks antigenic experience also of memory B cells. In mice, TFEB shapes the transcriptional landscape of germinal center B cells. Within the germinal center, TFEB facilitates the dark zone entry of light-zone-residing centrocytes through regulation of chemokine receptors and, by balancing the expression of Bcl-2/BH3-only family members, integrates antigen-induced apoptosis with T cell-provided CD40 survival signals. Thus, TFEB reprograms antigen-primed germinal center B cells for cell fate decisions.
© 2024. The Author(s).
Conflict of interest statement
A. Ballabio is co-founder of CASMA Therapeutics and an advisory board member of Next Generation Diagnostics and Avilar Therapeutics. All other authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- TRR 274, Project A08/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR 274, Project A08/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR 274/2 2024 - 408885537/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1328, Project A01 - 335447717/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR 274/2 2024 - 408885537/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1328, Project A01 - 335447717/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR 274/2 2024 - 408885537/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1328, Project A01 - 335447717/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Grant agreement No. 101021345 (T-Neuron)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- Grant agreement No. 101021345 (T-Neuron)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- Grant agreement No. 101021345 (T-Neuron)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

