The current landscape of spatial biomarkers for prediction of response to immune checkpoint inhibition
- PMID: 39138341
- PMCID: PMC11322473
- DOI: 10.1038/s41698-024-00671-1
The current landscape of spatial biomarkers for prediction of response to immune checkpoint inhibition
Abstract
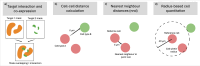
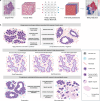
Enabling the examination of cell-cell relationships in tissue, spatially resolved omics technologies have revolutionised our perspectives on cancer biology. Clinically, the development of immune checkpoint inhibitors (ICI) has advanced cancer therapeutics. However, a major challenge of effective implementation is the identification of predictive biomarkers of response. In this review we examine the potential added predictive value of spatial biomarkers of response to ICI beyond current clinical benchmarks.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

