Critical review on in silico methods for structural annotation of chemicals detected with LC/HRMS non-targeted screening
- PMID: 39138659
- PMCID: PMC11700063
- DOI: 10.1007/s00216-024-05471-x
Critical review on in silico methods for structural annotation of chemicals detected with LC/HRMS non-targeted screening
Abstract
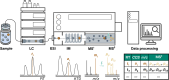
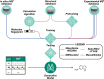
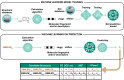
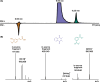
Non-targeted screening with liquid chromatography coupled to high-resolution mass spectrometry (LC/HRMS) is increasingly leveraging in silico methods, including machine learning, to obtain candidate structures for structural annotation of LC/HRMS features and their further prioritization. Candidate structures are commonly retrieved based on the tandem mass spectral information either from spectral or structural databases; however, the vast majority of the detected LC/HRMS features remain unannotated, constituting what we refer to as a part of the unknown chemical space. Recently, the exploration of this chemical space has become accessible through generative models. Furthermore, the evaluation of the candidate structures benefits from the complementary empirical analytical information such as retention time, collision cross section values, and ionization type. In this critical review, we provide an overview of the current approaches for retrieving and prioritizing candidate structures. These approaches come with their own set of advantages and limitations, as we showcase in the example of structural annotation of ten known and ten unknown LC/HRMS features. We emphasize that these limitations stem from both experimental and computational considerations. Finally, we highlight three key considerations for the future development of in silico methods.
Keywords: Generative modeling; Machine learning; Non-targeted analysis; Non-targeted screening; Suspect screening; Untargeted screening.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests.
Figures




Similar articles
-
Complementary methods for structural assignment of isomeric candidate structures in non-target liquid chromatography ion mobility high-resolution mass spectrometric analysis.Anal Bioanal Chem. 2023 Sep;415(21):5247-5259. doi: 10.1007/s00216-023-04852-y. Epub 2023 Jul 15. Anal Bioanal Chem. 2023. PMID: 37452839 Free PMC article.
-
[A novel method for efficient screening and annotation of important pathway-associated metabolites based on the modified metabolome and probe molecules].Se Pu. 2022 Sep;40(9):788-796. doi: 10.3724/SP.J.1123.2022.03025. Se Pu. 2022. PMID: 36156625 Free PMC article. Chinese.
-
Optimization of a liquid chromatography-ion mobility-high resolution mass spectrometry platform for untargeted lipidomics and application to HepaRG cell extracts.Talanta. 2021 Dec 1;235:122808. doi: 10.1016/j.talanta.2021.122808. Epub 2021 Aug 17. Talanta. 2021. PMID: 34517665
-
Non-targeted analysis (NTA) and suspect screening analysis (SSA): a review of examining the chemical exposome.J Expo Sci Environ Epidemiol. 2023 Jul;33(4):524-536. doi: 10.1038/s41370-023-00574-6. Epub 2023 Jun 28. J Expo Sci Environ Epidemiol. 2023. PMID: 37380877 Free PMC article. Review.
-
Recent advances in non-targeted screening analysis using liquid chromatography - high resolution mass spectrometry to explore new biomarkers for human exposure.Talanta. 2020 Nov 1;219:121339. doi: 10.1016/j.talanta.2020.121339. Epub 2020 Jul 7. Talanta. 2020. PMID: 32887069 Review.
Cited by
-
Do experimental projection methods outcompete retention time prediction models in non-target screening? A case study on LC/HRMS interlaboratory comparison data.Analyst. 2025 Aug 4;150(16):3567-3577. doi: 10.1039/d5an00323g. Analyst. 2025. PMID: 40671565 Free PMC article.
-
Assessing the Impact of Measurement Precision on Metabolite Identification Probability in Multidimensional Mass Spectrometry-Based, Reference-Free Metabolomics.Anal Chem. 2025 Jul 8;97(26):13861-13871. doi: 10.1021/acs.analchem.5c01067. Epub 2025 Jun 25. Anal Chem. 2025. PMID: 40556554 Free PMC article.
-
Large-scale generation of in silico based spectral libraries to annotate dark chemical space features in non-target analysis.Anal Bioanal Chem. 2025 Sep 2. doi: 10.1007/s00216-025-06034-4. Online ahead of print. Anal Bioanal Chem. 2025. PMID: 40892243
References
-
- Black G, Lowe C, Anumol T, Bade J, Favela K, Feng Y-L, Knolhoff A, Mceachran A, Nuñez J, Fisher C, Peter K, Quinete NS, Sobus J, Sussman E, Watson W, Wickramasekara S, Williams A, Young T. Exploring chemical space in non-targeted analysis: a proposed ChemSpace tool. Anal Bioanal Chem. 2023;415:35–44. 10.1007/s00216-022-04434-4. - PMC - PubMed
-
- Hollender J, Schymanski EL, Ahrens L, Alygizakis N, Béen F, Bijlsma L, Brunner AM, Celma A, Fildier A, Fu Q, Gago-Ferrero P, Gil-Solsona R, Haglund P, Hansen M, Kaserzon S, Kruve A, Lamoree M, Margoum C, Meijer J, Merel S, Rauert C, Rostkowski P, Samanipour S, Schulze B, Schulze T, Singh RR, Slobodnik J, Steininger-Mairinger T, Thomaidis NS, Togola A, Vorkamp K, Vulliet E, Zhu L, Krauss M. NORMAN guidance on suspect and non-target screening in environmental monitoring. Environ Sci Eur. 2023;35:75. 10.1186/s12302-023-00779-4.
-
- Manz KE, Feerick A, Braun JM, Feng Y-L, Hall A, Koelmel J, Manzano C, Newton SR, Pennell KD, Place BJ, Godri Pollitt KJ, Prasse C, Young JA. Non-targeted analysis (NTA) and suspect screening analysis (SSA): a review of examining the chemical exposome. J Expo Sci Environ Epidemiol. 2023;33:524–36. 10.1038/s41370-023-00574-6. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

