CD81 fusion alters SARS-CoV-2 Spike trafficking
- PMID: 39140770
- PMCID: PMC11389398
- DOI: 10.1128/mbio.01922-24
CD81 fusion alters SARS-CoV-2 Spike trafficking
Abstract
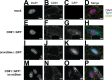
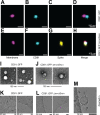
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic caused the biggest public health crises in recent history. Many expect future coronavirus introductions into the human population. Hence, it is essential to understand the basic biology of these viruses. In natural infection, the SARS-CoV-2 Spike (S) glycoprotein is co-expressed with all other viral proteins, which modify cellular compartments to maximize virion assembly. By comparison, most of S is degraded when the protein is expressed in isolation, as in current molecular vaccines. To probe the maturation pathway of S, we redirected its maturation by fusing S to the tetraspanin protein CD81. CD81 is a defining constituent of extracellular vesicles (EVs) or exosomes. EVs are generated in large numbers by all cells, extruded into blood and lymph, and transfer cargo between cells and systemically (estimated 1012 EVs per mL plasma). EVs, like platelets, can be transfused between unrelated donors. When fusing the proline-stabilized form of strain Delta S into the flexible, large extracellular loop of CD81 rather than being degraded in the lysosome, S was extruded into EVs. CD81-S fusion containing EVs were produced in large numbers and could be isolated to high purity. Purified CD81::S EVs bound ACE2, and S displayed on individual EV was observed by cryogenic electron microscopy (EM). The CD81::S-fusion EVs were non-toxic and elicited an anti-S trimer and anti-RBD antibody response in mice. This report shows a design path to maximize viral glycoprotein assembly and release without relying on the co-expression of potentially pathogenic nonstructural viral proteins.
Importance: The severe acute respiratory syndrome coronavirus 2 pandemic caused the biggest public health crises in recent history. To understand the maturation pathway of S, we fused S to the tetraspanin protein CD81. The resulting molecule is secreted in extracellular vesicles and induces antibodies in mice. This may be a general design path for viral glycoprotein vaccines.
Keywords: CD63; CD81; SARS-CoV-2; Spike; TSPAN; coronavirus; dSTORM; exosome; extracellular vesicles; tetraspanins.
Conflict of interest statement
D.P.D., A.S.C., R.P.M., and Y.Z. declare competing interests with respect to the possible commercialization (WO 2024/ 073397) of some of the information presented. These are managed by the University of North Carolina. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or conflict with the subject matter or materials discussed in the paper apart from those disclosed.
Figures



References
-
- Barouch DH, Stephenson KE, Sadoff J, Yu J, Chang A, Gebre M, McMahan K, Liu J, Chandrashekar A, Patel S, Le Gars M, de Groot AM, Heerwegh D, Struyf F, Douoguih M, van Hoof J, Schuitemaker H. 2021. Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N Engl J Med 385:951–953. doi: 10.1056/NEJMc2108829 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- P30 ES010126/ES/NIEHS NIH HHS/United States
- R01 DE018304/DE/NIDCR NIH HHS/United States
- T32 5T32AI007151/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 CA228172/CA/NCI NIH HHS/United States
- T32 2T32AI007419/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
