Transcription of HIV-1 at sites of intact latent provirus integration
- PMID: 39141127
- PMCID: PMC11323366
- DOI: 10.1084/jem.20240391
Transcription of HIV-1 at sites of intact latent provirus integration
Erratum in
-
Correction: Transcription of HIV-1 at sites of intact latent provirus integration.J Exp Med. 2025 Jun 2;222(6):e2024039105162025c. doi: 10.1084/jem.2024039105162025c. Epub 2025 May 23. J Exp Med. 2025. PMID: 40408268 Free PMC article. No abstract available.
Abstract
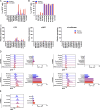
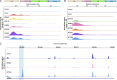
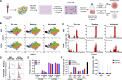
HIV-1 antiretroviral therapy is highly effective but fails to eliminate a reservoir of latent proviruses, leading to a requirement for life-long treatment. How the site of integration of authentic intact latent proviruses might impact their own or neighboring gene expression or reservoir dynamics is poorly understood. Here, we report on proviral and neighboring gene transcription at sites of intact latent HIV-1 integration in cultured T cells obtained directly from people living with HIV, as well as engineered primary T cells and cell lines. Proviral gene expression was correlated to the level of endogenous gene expression under resting but not activated conditions. Notably, latent proviral promoters were 100-10,000× less active than in productively infected cells and had little or no measurable impact on neighboring gene expression under resting or activated conditions. Thus, the site of integration has a dominant effect on the transcriptional activity of intact HIV-1 proviruses in the latent reservoir, thereby influencing cytopathic effects and proviral immune evasion.
© 2024 Teixeira et al.
Conflict of interest statement
Disclosures: R.B. Jones reports personal fees from ViiV Healthcare outside the submitted work. M.C Nussenzweig reports personal fees from Gilead and personal fees from Frontier Biosciences outside the submitted work; in addition, M.C. Nussenzweig has a patent to 3BNC117 issued (Gilead) and a patent to 10-1074 issued (Gilead). No other disclosures were reported.
Figures







Update of
-
Transcription of HIV-1 at sites of intact latent provirus integration.bioRxiv [Preprint]. 2024 Apr 29:2024.04.26.591331. doi: 10.1101/2024.04.26.591331. bioRxiv. 2024. Update in: J Exp Med. 2024 Sep 2;221(9):e20240391. doi: 10.1084/jem.20240391. PMID: 38746186 Free PMC article. Updated. Preprint.
References
-
- Antar, A.A.R., Jenike K.M., Jang S., Rigau D.N., Reeves D.B., Hoh R., Krone M.R., Keruly J.C., Moore R.D., Schiffer J.T., et al. 2020. Longitudinal study reveals HIV-1-infected CD4+ T cell dynamics during long-term antiretroviral therapy. J. Clin. Invest. 130:3543–3559. 10.1172/JCI135953 - DOI - PMC - PubMed
-
- Battivelli, E., Dahabieh M.S., Abdel-Mohsen M., Svensson J.P., Tojal Da Silva I., Cohn L.B., Gramatica A., Deeks S., Greene W.C., Pillai S.K., and Verdin E.. 2018. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4(+) T cells. Elife. 7:e34655. 10.7554/eLife.34655 - DOI - PMC - PubMed
-
- Bui, J.K., Sobolewski M.D., Keele B.F., Spindler J., Musick A., Wiegand A., Luke B.T., Shao W., Hughes S.H., Coffin J.M., et al. 2017. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13:e1006283. 10.1371/journal.ppat.1006283 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

