mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR
- PMID: 39141345
- PMCID: PMC11348030
- DOI: 10.1073/pnas.2405959121
mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR
Abstract
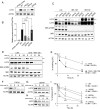
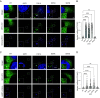
TORC1 (target of rapamycin complex 1) is a highly conserved protein kinase that plays a central role in regulating cell growth. Given the role of mammalian TORC1 (mTORC1) in metabolism and disease, understanding mTORC1 downstream signaling and feedback loops is important. mTORC1 recognizes some of its substrates via a five amino acid binding sequence called the TOR signaling (TOS) motif. mTORC1 binding to a TOS motif facilitates phosphorylation of a distinct, distal site. Here, we show that LST2, also known as ZFYVE28, contains a TOS motif (amino acids 401 to 405) and is directly phosphorylated by mTORC1 at serine 670 (S670). mTORC1-mediated S670 phosphorylation promotes LST2 monoubiquitination on lysine 87 (K87). Monoubiquitinated LST2 is stable and displays a broad reticular distribution. When mTORC1 is inactive, unphosphorylated LST2 is degraded by the proteasome. The absence of LST2 enhances EGFR (epidermal growth factor receptor) signaling. We propose that mTORC1 negatively feeds back on its upstream receptor EGFR via LST2.
Keywords: LST2; TOS motif; mTOR signaling; negative feedback; phosphorylation substrate.
Conflict of interest statement
Competing interests statement:T.M. and M.N.H. collaborate with Aukera Therapeutics GmbH. M.N.H. is affiliated with F. Hoffmann-La Roche Ltd., Swiss Rockets AG and Torqur AG.
Figures



References
-
- Dazert E., Hall M. N., mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 (2011). - PubMed
-
- Battaglioni S., Benjamin D., Wälchli M., Maier T., Hall M. N., mTOR substrate phosphorylation in growth control. Cell 185, 1814–1836 (2022). - PubMed
-
- Loewith R., et al. , Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 (2002). - PubMed
-
- Wullschleger S., Loewith R., Hall M. N., TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- 205601/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)
- NA/Sjöbergstiftelsen (Sjöberg Foundation)
- NA/Louis-Jeantet Foundation (Fondation Louis-Jeantet)
- 812830/H2020 ITN Healthage
- 179323/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

