TET3-overexpressing macrophages promote endometriosis
- PMID: 39141428
- PMCID: PMC11527447
- DOI: 10.1172/JCI181839
TET3-overexpressing macrophages promote endometriosis
Abstract
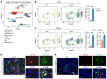
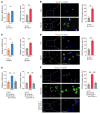
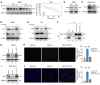
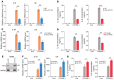
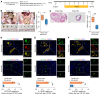
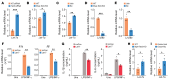
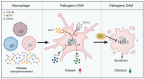
Endometriosis is a debilitating, chronic inflammatory disease affecting approximately 10% of reproductive-age women worldwide with no cure. While macrophages have been intrinsically linked to the pathophysiology of endometriosis, targeting them therapeutically has been extremely challenging due to their high heterogeneity and because these disease-associated macrophages (DAMs) can be either pathogenic or protective. Here, we report identification of pathogenic macrophages characterized by TET3 overexpression in human endometriosis lesions. We show that factors from the disease microenvironment upregulated TET3 expression, transforming macrophages into pathogenic DAMs. TET3 overexpression stimulated proinflammatory cytokine production via a feedback mechanism involving inhibition of let-7 miRNA expression. Remarkably, these cells relied on TET3 overexpression for survival and hence were vulnerable to TET3 knockdown. We demonstrated that Bobcat339, a synthetic cytosine derivative, triggered TET3 degradation in both human and mouse macrophages. This degradation was dependent on a von Hippel-Lindau (VHL) E3 ubiquitin ligase whose expression was also upregulated in TET3-overexpressing macrophages. Furthermore, depleting TET3-overexpressing macrophages either through myeloid-specific Tet3 ablation or using Bobcat339 strongly inhibited endometriosis progression in mice. Our results defined TET3-overexpressing macrophages as key pathogenic contributors to and attractive therapeutic targets for endometriosis. Our findings may also be applicable to other chronic inflammatory diseases where DAMs have important roles.
Keywords: Inflammation; Macrophages; Obstetrics/gynecology; Pain; Reproductive biology.
Figures


Comment in
- Targeting TET3 in macrophages provides a concept strategy for the treatment of endometriosis doi: 10.1172/JCI185421
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

