Aging human abdominal subcutaneous white adipose tissue at single cell resolution
- PMID: 39141531
- PMCID: PMC11561672
- DOI: 10.1111/acel.14287
Aging human abdominal subcutaneous white adipose tissue at single cell resolution
Abstract
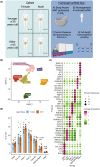
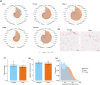
White adipose tissue (WAT) is a robust energy storage and endocrine organ critical for maintaining metabolic health as we age. Our aim was to identify cell-specific transcriptional aberrations that occur in WAT with aging. We leveraged full-length snRNA-Seq and histology to characterize the cellular landscape of human abdominal subcutaneous WAT in a prospective cohort of 10 younger (≤30 years) and 10 older individuals (≥65 years) balanced for sex and body mass index (BMI). The older group had greater cholesterol, very-low-density lipoprotein, triglycerides, thyroid stimulating hormone, and aspartate transaminase compared to the younger group (p < 0.05). We highlight that aging WAT is associated with adipocyte hypertrophy, increased proportions of lipid-associated macrophages and mast cells, an upregulation of immune responses linked to fibrosis in pre-adipocyte, adipocyte, and vascular populations, and highlight CXCL14 as a biomarker of these processes. We show that older WAT has elevated levels of senescence marker p16 in adipocytes and identify the adipocyte subpopulation driving this senescence profile. We confirm that these transcriptional and phenotypical changes occur without overt fibrosis and in older individuals that have comparable WAT insulin sensitivity to the younger individuals.
Keywords: White adipose tissue; aging; senescence; single nuclei RNA‐Seq.
© 2024 The Author(s). Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Update of
-
A single nuclei atlas of aging human abdominal subcutaneous white adipose tissue.Res Sq [Preprint]. 2023 Jul 13:rs.3.rs-3097605. doi: 10.21203/rs.3.rs-3097605/v1. Res Sq. 2023. Update in: Aging Cell. 2024 Nov;23(11):e14287. doi: 10.1111/acel.14287. PMID: 37503028 Free PMC article. Updated. Preprint.
References
-
- Acosta, J. R. , Joost, S. , Karlsson, K. , Ehrlund, A. , Li, X. , Aouadi, M. , Kasper, M. , Arner, P. , Rydén, M. , & Laurencikiene, J. (2017). Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Research & Therapy, 8(1), 250. 10.1186/s13287-017-0701-4 - DOI - PMC - PubMed
-
- Angueira, A. R. , Sakers, A. P. , Holman, C. D. , Cheng, L. , Arbocco, M. N. , Shamsi, F. , Lynes, M. D. , Shrestha, R. , Okada, C. , Batmanov, K. , Susztak, K. , Tseng, Y. H. , Liaw, L. , & Seale, P. (2021). Defining the lineage of thermogenic perivascular adipose tissue. Nature Metabolism, 3(4), 469–484. 10.1038/s42255-021-00380-0 - DOI - PMC - PubMed
-
- Aversa, Z. , White, T. A. , Heeren, A. A. , Hulshizer, C. A. , Saul, D. , Zhang, X. , Molina, A. J. A. , Redman, L. M. , Martin, C. K. , Racette, S. B. , Huffman, K. M. , Bhapkar, M. , Khosla, S. , Das, S. K. , Fielding, R. A. , Atkinson, E. J. , & LeBrasseur, N. K. (2023). Calorie restriction reduces biomarkers of cellular senescence in humans. Aging Cell, 23, e14038. 10.1111/acel.14038 - DOI - PMC - PubMed
-
- Bäckdahl, J. , Franzén, L. , Massier, L. , Li, Q. , Jalkanen, J. , Gao, H. , Andersson, A. , Bhalla, N. , Thorell, A. , Rydén, M. , Ståhl, P. L. , & Mejhert, N. (2021). Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin. Cell Metabolism, 33, 1869–1882. 10.1016/j.cmet.2021.07.018.e6. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

