Risk Variants Associated With Normal Pressure Hydrocephalus: Genome-Wide Association Study in the FinnGen Cohort
- PMID: 39141892
- PMCID: PMC11361830
- DOI: 10.1212/WNL.0000000000209694
Risk Variants Associated With Normal Pressure Hydrocephalus: Genome-Wide Association Study in the FinnGen Cohort
Abstract
Background and objectives: Large-scale genome-wide studies of chronic hydrocephalus have been lacking. We conducted a genome-wide association study (GWAS) in normal pressure hydrocephalus (NPH).
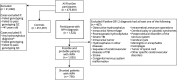
Methods: We used a case-control study design implementing FinnGen data containing 473,691 Finns with genotypes and nationwide health records. Patients with NPH were selected based on ICD-10 G91.2 diagnosis. To select patients with idiopathic NPH (iNPH) for sensitivity analysis, we excluded patients with a potentially known etiology of the condition using an algorithm on their disease history. The controls were the remaining non-hydrocephalic participants. For a replication analysis, the NPH cohort from UK Biobank (UKBB) was used.
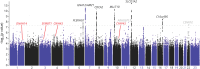
Results: We included 1,522 patients with NPH (mean age 72.2 years, 53% women) and 451,091 controls (mean age 60.5 years, 44% women). In the GWAS comparing patients with NPH with the controls, we identified 6 gene regions significantly (p < 5.0e-8) associated with NPH that replicated in a meta-analysis with UKBB (NPH n = 173). The top loci near the following genes were rs7962263, SLCO1A2 (odds ratio [OR] 0.71, 95% CI 0.65-0.78, p = 1.0e-14); rs798495, AMZ1/GNA12 (OR 1.29, 95% CI 1.20-1.39, p = 2.9e-12); rs10828247, MLLT10 (OR 0.77, 95% CI 0.71-0.83, p = 1.5e-11); rs561699566 and rs371919113, CDCA2 (OR 0.76, 95% CI 0.70-0.82, p = 1.5e-11); rs56023709, C16orf95 (OR 1.24, 95% CI 1.16-1.33, p = 3.0e-9); and rs62434144, PLEKHG1 (OR 1.23, 95% CI 1.14-1.32, p = 1.4e-8). In the sensitivity analysis comparing only patients with iNPH (n = 1,055) with the controls (n = 451,091), 4 top loci near the following genes remained significant: rs7962263, SLCO1A2 (OR 0.70, 95% CI 0.63-0.78, p = 2.1e-11); rs10828247, MLLT10 (OR 0.74, 95% CI 0.62-0.82, p = 4.6e-10); rs798511, AMZ1/GNA12 (OR 1.28, 95% CI 1.17-1.39, p = 1.7e-8); and rs56023709, C16orf95 (OR 1.28, 95% CI 1.17-1.39, p = 1.7e-8).
Discussion: We identified 6 loci significantly associated with NPH in the thus far largest GWAS in chronic hydrocephalus. The genes near the top loci have previously been associated with blood-brain barrier and blood-CSF barrier function and with increased lateral brain ventricle volume. The effect sizes and allele frequencies remained similar in NPH and iNPH cohorts, indicating the identified loci are risk determinants for iNPH and likely not explained by associations with other etiologies. However, the exact role of these loci is still unknown, warranting further studies.
Conflict of interest statement
The authors report no relevant disclosures. Go to
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
