Schwann cell-secreted PGE2 promotes sensory neuron excitability during development
- PMID: 39142281
- PMCID: PMC11967275
- DOI: 10.1016/j.cell.2024.07.033
Schwann cell-secreted PGE2 promotes sensory neuron excitability during development
Abstract
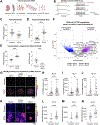
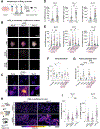
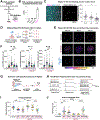
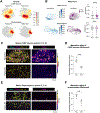
Electrical excitability-the ability to fire and propagate action potentials-is a signature feature of neurons. How neurons become excitable during development and whether excitability is an intrinsic property of neurons remain unclear. Here, we demonstrate that Schwann cells, the most abundant glia in the peripheral nervous system, promote somatosensory neuron excitability during development. We find that Schwann cells secrete prostaglandin E2, which is necessary and sufficient to induce developing somatosensory neurons to express normal levels of genes required for neuronal function, including voltage-gated sodium channels, and to fire action potential trains. Inactivating this signaling pathway in Schwann cells impairs somatosensory neuron maturation, causing multimodal sensory defects that persist into adulthood. Collectively, our studies uncover a neurodevelopmental role for prostaglandin E2 distinct from its established role in inflammation, revealing a cell non-autonomous mechanism by which glia regulate neuronal excitability to enable the development of normal sensory functions.
Keywords: PGE2; Schwann cells; excitability; glial cells; neural development; neuron-glia interactions; prostaglandin; sensory behavior; sensory neuron; voltage-gated sodium channels.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.D.B. is a cofounder and holds equity shares in SiteOne Therapeutics, Inc., a start-up company developing subtype-selective modulators of Na(V)s.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

