Genome-wide Studies Reveal Genetic Risk Factors for Hepatic Fat Content
- PMID: 39142818
- PMCID: PMC12016563
- DOI: 10.1093/gpbjnl/qzae031
Genome-wide Studies Reveal Genetic Risk Factors for Hepatic Fat Content
Abstract
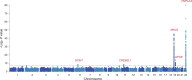
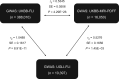
Genetic susceptibility to metabolic associated fatty liver disease (MAFLD) is complex and poorly characterized. Accurate characterization of the genetic background of hepatic fat content would provide insights into disease etiology and causality of risk factors. We performed genome-wide association study (GWAS) on two noninvasive definitions of hepatic fat content: magnetic resonance imaging proton density fat fraction (MRI-PDFF) in 16,050 participants and fatty liver index (FLI) in 388,701 participants from the United Kingdom (UK) Biobank (UKBB). Heritability, genetic overlap, and similarity between hepatic fat content phenotypes were analyzed, and replicated in 10,398 participants from the University Medical Center Groningen (UMCG) Genetics Lifelines Initiative (UGLI). Meta-analysis of GWASs of MRI-PDFF in UKBB revealed five statistically significant loci, including two novel genomic loci harboring CREB3L1 (rs72910057-T, P = 5.40E-09) and GCM1 (rs1491489378-T, P = 3.16E-09), respectively, as well as three previously reported loci: PNPLA3, TM6SF2, and APOE. GWAS of FLI in UKBB identified 196 genome-wide significant loci, of which 49 were replicated in UGLI, with top signals in ZPR1 (P = 3.35E-13) and FTO (P = 2.11E-09). Statistically significant genetic correlation (rg) between MRI-PDFF (UKBB) and FLI (UGLI) GWAS results was found (rg = 0.5276, P = 1.45E-03). Novel MRI-PDFF genetic signals (CREB3L1 and GCM1) were replicated in the FLI GWAS. We identified two novel genes for MRI-PDFF and 49 replicable loci for FLI. Despite a difference in hepatic fat content assessment between MRI-PDFF and FLI, a substantial similar genetic architecture was found. FLI is identified as an easy and reliable approach to study hepatic fat content at the population level.
Keywords: Fatty liver index; Genome-wide association study; Hepatic fat content; MAFLD; Magnetic resonance imaging proton density fat fraction.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
Rinse K. Weersma has received unrestricted research grants from Takeda, Johnson & Johnson, Ferring, and Tramedico as well as speaker fees from AbbVie, MSD, and Boston Scientific, and has acted as a consultant for Takeda Pharmaceuticals. All the other authors have declared no competing interests.
Figures

References
-
- Arab JP, Arrese M, Trauner M.. Recent insights into the pathogenesis of nonalcoholic fatty liver disease. Annu Rev Pathol 2018;13:321–50. - PubMed
-
- Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, et al.A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J Hepatol 2020;73:202–9. - PubMed
-
- Anstee QM, Darlay R, Cockell S, Meroni M, Govaere O, Tiniakos D, et al.Genome-wide association study of non-alcoholic fatty liver and steatohepatitis in a histologically characterised cohort. J Hepatol 2020;73:505–15. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

