Metabolic alterations in vitamin D deficient systemic lupus erythematosus patients
- PMID: 39143130
- PMCID: PMC11325032
- DOI: 10.1038/s41598-024-67588-4
Metabolic alterations in vitamin D deficient systemic lupus erythematosus patients
Abstract
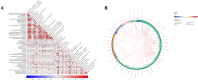
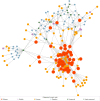
Vitamin D deficiency is increasingly common in systemic lupus erythematosus (SLE) patients and is associated with the disease activity and proteinuria. Recently, alterations in metabolism have been recognized as key regulators of SLE pathogenesis. Our objective was to identify differential metabolites in the serum metabolome of SLE with vitamin D deficiency. In this study, serum samples from 31 SLE patients were collected. Levels of 25(OH)D3 were assayed by ELISA. Patients were divided into two groups according to their vitamin D level (20 ng/ml). Untargeted metabolomics were used to study the metabolite profiles in serum by high-performance liquid chromatography-tandem mass spectrometry. Subsequently, we performed metabolomics profiling analysis to identify 52 significantly altered metabolites in vitamin D deficient SLE patients. The area under the curve (AUC) from ROC analyses was calculated to assess the diagnostic potential of each candidate metabolite biomarker. Lipids accounted for 66.67% of the differential metabolites in the serum, highlighted the disruption of lipid metabolism. The 52 differential metabolites were mapped to 27 metabolic pathways, with fat digestion and absorption, as well as lipid metabolism, emerging as the most significant pathways. The AUC of (S)-Oleuropeic acid and 2-Hydroxylinolenic acid during ROC analysis were 0.867 and 0.833, respectively, indicating their promising diagnostic potential. In conclusion, our results revealed vitamin D deficiency alters SLE metabolome, impacting lipid metabolism, and thrown insights into the pathogenesis and diagnosis of SLE.
Keywords: Lipids; Metabolism; Systemic lupus erythematosus; Vitamin D.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

