Human organoids with an autologous tissue-resident immune compartment
- PMID: 39143209
- PMCID: PMC11374719
- DOI: 10.1038/s41586-024-07791-5
Human organoids with an autologous tissue-resident immune compartment
Abstract
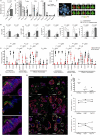
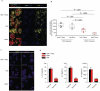
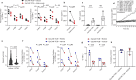
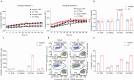
The intimate relationship between the epithelium and immune system is crucial for maintaining tissue homeostasis, with perturbations therein linked to autoimmune disease and cancer1-3. Whereas stem cell-derived organoids are powerful models of epithelial function4, they lack tissue-resident immune cells that are essential for capturing organ-level processes. We describe human intestinal immuno-organoids (IIOs), formed through self-organization of epithelial organoids and autologous tissue-resident memory T (TRM) cells, a portion of which integrate within the epithelium and continuously survey the barrier. TRM cell migration and interaction with epithelial cells was orchestrated by TRM cell-enriched transcriptomic programs governing cell motility and adhesion. We combined IIOs and single-cell transcriptomics to investigate intestinal inflammation triggered by cancer-targeting biologics in patients. Inflammation was associated with the emergence of an activated population of CD8+ T cells that progressively acquired intraepithelial and cytotoxic features. The appearance of this effector population was preceded and potentiated by a T helper-1-like CD4+ population, which initially produced cytokines and subsequently became cytotoxic itself. As a system amenable to direct perturbation, IIOs allowed us to identify the Rho pathway as a new target for mitigation of immunotherapy-associated intestinal inflammation. Given that they recapitulate both the phenotypic outcomes and underlying interlineage immune interactions, IIOs can be used to study tissue-resident immune responses in the context of tumorigenesis and infectious and autoimmune diseases.
© 2024. The Author(s).
Conflict of interest statement
All authors are employees of Hoffmann-LaRoche. The company provided support in the form of salaries for authors but did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Figures







References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

