Mitochondrial complex I promotes kidney cancer metastasis
- PMID: 39143213
- PMCID: PMC11424252
- DOI: 10.1038/s41586-024-07812-3
Mitochondrial complex I promotes kidney cancer metastasis
Abstract
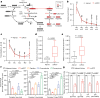
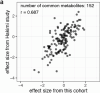
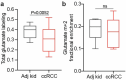
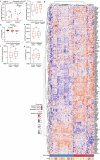
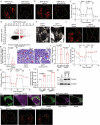
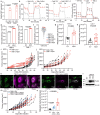
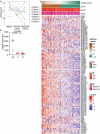
Most kidney cancers are metabolically dysfunctional1-4, but how this dysfunction affects cancer progression in humans is unknown. We infused 13C-labelled nutrients in over 80 patients with kidney cancer during surgical tumour resection. Labelling from [U-13C]glucose varies across subtypes, indicating that the kidney environment alone cannot account for all tumour metabolic reprogramming. Compared with the adjacent kidney, clear cell renal cell carcinomas (ccRCCs) display suppressed labelling of tricarboxylic acid (TCA) cycle intermediates in vivo and in ex vivo organotypic cultures, indicating that suppressed labelling is tissue intrinsic. [1,2-13C]acetate and [U-13C]glutamine infusions in patients, coupled with measurements of respiration in isolated human kidney and tumour mitochondria, reveal lower electron transport chain activity in ccRCCs that contributes to decreased oxidative and enhanced reductive TCA cycle labelling. However, ccRCC metastases unexpectedly have enhanced TCA cycle labelling compared with that of primary ccRCCs, indicating a divergent metabolic program during metastasis in patients. In mice, stimulating respiration or NADH recycling in kidney cancer cells is sufficient to promote metastasis, whereas inhibiting electron transport chain complex I decreases metastasis. These findings in humans and mice indicate that metabolic properties and liabilities evolve during kidney cancer progression, and that mitochondrial function is limiting for metastasis but not growth at the original site.
© 2024. The Author(s).
Conflict of interest statement
R.J.D. is a founder and advisor at Atavistik Bio, and serves on the Scientific Advisory Boards of Agios Pharmaceuticals, Vida Ventures and Droia Ventures. I.P. has served on Scientific Advisory Boards of Health Tech International, Merck and Otsuka, and is co-inventor of patents with Philips Healthcare. K.D.C. participated on an Advisory Board for Novartis. G.G. serves on the Advisory Board of Medendi.
Figures








Update of
-
Mitochondrial metabolism in primary and metastatic human kidney cancers.bioRxiv [Preprint]. 2023 Feb 7:2023.02.06.527285. doi: 10.1101/2023.02.06.527285. bioRxiv. 2023. Update in: Nature. 2024 Sep;633(8031):923-931. doi: 10.1038/s41586-024-07812-3. PMID: 36798172 Free PMC article. Updated. Preprint.
Comment in
-
Metabolism, mitochondria and metastasis in kidney cancer.Nat Rev Urol. 2024 Dec;21(12):705. doi: 10.1038/s41585-024-00968-7. Nat Rev Urol. 2024. PMID: 39537998 No abstract available.
References
MeSH terms
Substances
Grants and funding
- F30 CA254150/CA/NCI NIH HHS/United States
- F31 CA239330/CA/NCI NIH HHS/United States
- F31 DK122676/DK/NIDDK NIH HHS/United States
- U2C DK119889/DK/NIDDK NIH HHS/United States
- R35 CA220449/CA/NCI NIH HHS/United States
- P50 CA196516/CA/NCI NIH HHS/United States
- TL1 TR001104/TR/NCATS NIH HHS/United States
- T32 GM008203/GM/NIGMS NIH HHS/United States
- U2C DK119886/DK/NIDDK NIH HHS/United States
- U01 CA207091/CA/NCI NIH HHS/United States
- UL1 TR003163/TR/NCATS NIH HHS/United States
- R01 CA154475/CA/NCI NIH HHS/United States
- R00 CA248838/CA/NCI NIH HHS/United States
- P41 EB015908/EB/NIBIB NIH HHS/United States
- P30 CA142543/CA/NCI NIH HHS/United States
- OT2 OD030544/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

