Spatial control of the APC/C ensures the rapid degradation of cyclin B1
- PMID: 39143240
- PMCID: PMC11445581
- DOI: 10.1038/s44318-024-00194-2
Spatial control of the APC/C ensures the rapid degradation of cyclin B1
Abstract
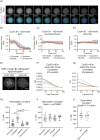
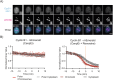
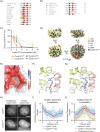
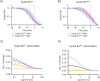
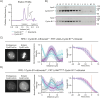
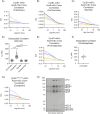
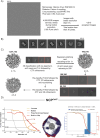
The proper control of mitosis depends on the ubiquitin-mediated degradation of the right mitotic regulator at the right time. This is effected by the Anaphase Promoting Complex/Cyclosome (APC/C) ubiquitin ligase that is regulated by the Spindle Assembly Checkpoint (SAC). The SAC prevents the APC/C from recognising Cyclin B1, the essential anaphase and cytokinesis inhibitor, until all chromosomes are attached to the spindle. Once chromosomes are attached, Cyclin B1 is rapidly degraded to enable chromosome segregation and cytokinesis. We have a good understanding of how the SAC inhibits the APC/C, but relatively little is known about how the APC/C recognises Cyclin B1 as soon as the SAC is turned off. Here, by combining live-cell imaging, in vitro reconstitution biochemistry, and structural analysis by cryo-electron microscopy, we provide evidence that the rapid recognition of Cyclin B1 in metaphase requires spatial regulation of the APC/C. Using fluorescence cross-correlation spectroscopy, we find that Cyclin B1 and the APC/C primarily interact at the mitotic apparatus. We show that this is because Cyclin B1, like the APC/C, binds to nucleosomes, and identify an 'arginine-anchor' in the N-terminus as necessary and sufficient for binding to the nucleosome. Mutating the arginine anchor on Cyclin B1 reduces its interaction with the APC/C and delays its degradation: cells with the mutant, non-nucleosome-binding Cyclin B1 become aneuploid, demonstrating the physiological relevance of our findings. Together, our data demonstrate that mitotic chromosomes promote the efficient interaction between Cyclin B1 and the APC/C to ensure the timely degradation of Cyclin B1 and genomic stability.
Keywords: Cell Cycle; Chromosome; Mitosis; Nucleosome; Ubiquitin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Acquaviva C, Pines J (2006) The anaphase-promoting complex/cyclosome: APC/C. J Cell Sci 119:2401–2404 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

