Identification and analysis of inflammation-related biomarkers in tetralogy of Fallot
- PMID: 39144431
- PMCID: PMC11320004
- DOI: 10.21037/tp-24-8
Identification and analysis of inflammation-related biomarkers in tetralogy of Fallot
Abstract
Background: Studies have revealed that inflammatory response is relevant to the tetralogy of Fallot (TOF). However, there are no studies to systematically explore the role of the inflammation-related genes (IRGs) in TOF. Therefore, based on bioinformatics, we explored the biomarkers related to inflammation in TOF, laying a theoretical foundation for its in-depth study.
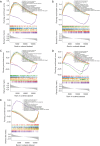
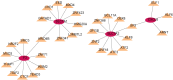
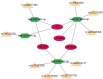
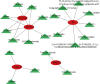
Methods: TOF-related datasets (GSE36761 and GSE35776) were downloaded from the Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) between TOF and control groups were identified in GSE36761. And DEGs between TOF and control groups were intersected with IRGs to obtain differentially expressed IRGs (DE-IRGs). Afterwards, the least absolute shrinkage and selection operator (LASSO) and random forest (RF) were utilized to identify the biomarkers. Next, immune analysis was carried out. The transcription factor (TF)-mRNA, lncRNA-miRNA-mRNA, and miRNA-single nucleotide polymorphism (SNP)-mRNA networks were created. Finally, the potential drugs targeting the biomarkers were predicted.
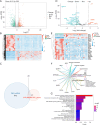
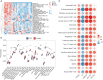
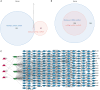
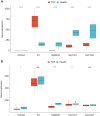
Results: There were 971 DEGs between TOF and control groups, and 29 DE-IRGs were gained through the intersection between DEGs and IRGs. Next, a total of five biomarkers (MARCO, CXCL6, F3, SLC7A2, and SLC7A1) were acquired via two machine learning algorithms. Infiltrating abundance of 18 immune cells was significantly different between TOF and control groups, such as activated B cells, neutrophil, CD56dim natural killer cells, etc. The TF-mRNA network contained 4 mRNAs, 31 TFs, and 33 edges, for instance, ELF1-CXCL6, CBX8-SLC7A2, ZNF423-SLC7A1, ZNF71-F3. The lncRNA-miRNA-mRNA network was created, containing 4 mRNAs, 4 miRNAs, and 228 lncRNAs. Afterwards, nine SNPs locations were identified in the miRNA-SNP-mRNA network. A total of 21 drugs were predicted, such as ornithine, lysine, arginine, etc.
Conclusions: Our findings detected five inflammation-related biomarkers (MARCO, CXCL6, F3, SLC7A2, and SLC7A1) for TOF, providing a scientific reference for further studies of TOF.
Keywords: Tetralogy of Fallot (TOF); bioinformatics analysis; biomarkers; inflammation; network.
2024 Translational Pediatrics. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-24-8/coif). The authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
