Navigating fluoroquinolone resistance in Gram-negative bacteria: a comprehensive evaluation
- PMID: 39144447
- PMCID: PMC11323783
- DOI: 10.1093/jacamr/dlae127
Navigating fluoroquinolone resistance in Gram-negative bacteria: a comprehensive evaluation
Abstract
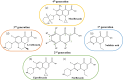
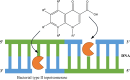
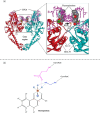
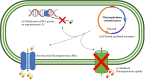
Since the introduction of quinolone and fluoroquinolone antibiotics to treat bacterial infections in the 1960s, there has been a pronounced increase in the number of bacterial species that have developed resistance to fluoroquinolone treatment. In 2017, the World Health Organization established a priority list of the most critical Gram-negative resistant pathogens. These included Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli. In the last three decades, investigations into the mechanisms of fluoroquinolone resistance have revealed that mutations in the target enzymes of fluoroquinolones, DNA gyrase or topoisomerase IV, are the most prevalent mechanism conferring high levels of resistance. Alterations to porins and efflux pumps that facilitate fluoroquinolone permeation and extrusion across the bacterial cell membrane also contribute to the development of resistance. However, there is a growing observation of novel mutants with newer generations of fluoroquinolones, highlighting the need for novel treatments. Currently, steady progress has been made in the development of novel antimicrobial agents that target DNA gyrase or topoisomerase IV through different avenues than current fluoroquinolones to prevent target-mediated resistance. Therefore, an updated review of the current understanding of fluoroquinolone resistance within the literature is imperative to aid in future investigations.
© The Author(s) 2024. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures
Similar articles
-
Analysis of mutational patterns in quinolone resistance-determining regions of GyrA and ParC of clinical isolates.Int J Antimicrob Agents. 2019 Mar;53(3):318-324. doi: 10.1016/j.ijantimicag.2018.12.004. Epub 2018 Dec 21. Int J Antimicrob Agents. 2019. PMID: 30582984
-
Gyrase and Topoisomerase IV: Recycling Old Targets for New Antibacterials to Combat Fluoroquinolone Resistance.ACS Infect Dis. 2024 Apr 12;10(4):1097-1115. doi: 10.1021/acsinfecdis.4c00128. Epub 2024 Apr 2. ACS Infect Dis. 2024. PMID: 38564341 Free PMC article. Review.
-
In vitro activity of WQ-3810, a novel fluoroquinolone, against multidrug-resistant and fluoroquinolone-resistant pathogens.Int J Antimicrob Agents. 2014 Nov;44(5):443-9. doi: 10.1016/j.ijantimicag.2014.07.017. Epub 2014 Sep 1. Int J Antimicrob Agents. 2014. PMID: 25239276
-
The Landscape of Phenotypic and Transcriptional Responses to Ciprofloxacin in Acinetobacter baumannii: Acquired Resistance Alleles Modulate Drug-Induced SOS Response and Prophage Replication.mBio. 2019 Jun 11;10(3):e01127-19. doi: 10.1128/mBio.01127-19. mBio. 2019. PMID: 31186328 Free PMC article.
-
Sulopenem: An Intravenous and Oral Penem for the Treatment of Urinary Tract Infections Due to Multidrug-Resistant Bacteria.Drugs. 2022 Apr;82(5):533-557. doi: 10.1007/s40265-022-01688-1. Epub 2022 Mar 16. Drugs. 2022. PMID: 35294769 Review.
Cited by
-
Drug susceptibility of uropathogens isolated from patients treated at the Mazovian Specialized Hospital in Radom.Acta Biochim Pol. 2025 Feb 27;72:14082. doi: 10.3389/abp.2025.14082. eCollection 2025. Acta Biochim Pol. 2025. PMID: 40083641 Free PMC article.
-
War and peace: exploring microbial defence systems as a source of new antimicrobial therapies.Front Pharmacol. 2025 Jan 7;15:1504901. doi: 10.3389/fphar.2024.1504901. eCollection 2024. Front Pharmacol. 2025. PMID: 39840088 Free PMC article. Review.
-
Facing antimicrobial resistance in cancer care: what the AIOM survey tells us about oncologists' awareness.JAC Antimicrob Resist. 2025 Aug 18;7(4):dlaf150. doi: 10.1093/jacamr/dlaf150. eCollection 2025 Aug. JAC Antimicrob Resist. 2025. PMID: 40831750 Free PMC article.
-
Urinary Tract Infections Caused by Klebsiella pneumoniae and Prolonged Treatment with Trimethoprim/Sulfamethoxazole.Microorganisms. 2025 Feb 14;13(2):422. doi: 10.3390/microorganisms13020422. Microorganisms. 2025. PMID: 40005786 Free PMC article.
-
The Rise, Fall, and Rethink of (Fluoro)quinolones: A Quick Rundown.Pathogens. 2025 May 24;14(6):525. doi: 10.3390/pathogens14060525. Pathogens. 2025. PMID: 40559533 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
