Assessing patterns of metazoans in the global ocean using environmental DNA
- PMID: 39144493
- PMCID: PMC11321857
- DOI: 10.1098/rsos.240724
Assessing patterns of metazoans in the global ocean using environmental DNA
Abstract
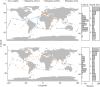
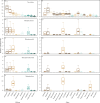
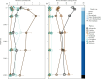
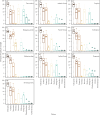
Documenting large-scale patterns of animals in the ocean and determining the drivers of these patterns is needed for conservation efforts given the unprecedented rates of change occurring within marine ecosystems. We used existing datasets from two global expeditions, Tara Oceans and Malaspina, that circumnavigated the oceans and sampled down to 4000 m to assess metazoans from environmental DNA (eDNA) extracted from seawater. We describe patterns of taxonomic richness within metazoan phyla and orders based on metabarcoding and infer the relative abundance of phyla using metagenome datasets, and relate these data to environmental variables. Arthropods had the greatest taxonomic richness of metazoan phyla at the surface, while cnidarians had the greatest richness in pelagic zones. Half of the marine metazoan eDNA from metagenome datasets was from arthropods, followed by cnidarians and nematodes. We found that mean surface temperature and primary productivity were positively related to metazoan taxonomic richness. Our findings concur with existing knowledge that temperature and primary productivity are important drivers of taxonomic richness for specific taxa at the ocean's surface, but these correlations are less evident in the deep ocean. Massive sequencing of eDNA can improve understanding of animal distributions, particularly for the deep ocean where sampling is challenging.
Keywords: deep sea; diversity; global abundance; metabarcoding; metagenome; metazoan.
© 2024 The Authors.
Conflict of interest statement
We declare we have no competing interests.
Figures
Similar articles
-
Capturing open ocean biodiversity: Comparing environmental DNA metabarcoding to the continuous plankton recorder.Mol Ecol. 2021 Jul;30(13):3140-3157. doi: 10.1111/mec.15587. Epub 2020 Sep 1. Mol Ecol. 2021. PMID: 32767849
-
Hydroids (Cnidaria, Hydrozoa) from Mauritanian Coral Mounds.Zootaxa. 2020 Nov 16;4878(3):zootaxa.4878.3.2. doi: 10.11646/zootaxa.4878.3.2. Zootaxa. 2020. PMID: 33311142
-
Environmental DNA surveys detect distinct metazoan communities across abyssal plains and seamounts in the western Clarion Clipperton Zone.Mol Ecol. 2020 Dec;29(23):4588-4604. doi: 10.1111/mec.15484. Epub 2020 Jun 14. Mol Ecol. 2020. PMID: 32452072 Free PMC article.
-
The Minderoo-Monaco Commission on Plastics and Human Health.Ann Glob Health. 2023 Mar 21;89(1):23. doi: 10.5334/aogh.4056. eCollection 2023. Ann Glob Health. 2023. PMID: 36969097 Free PMC article. Review.
-
Environmental DNA metabarcoding: Transforming how we survey animal and plant communities.Mol Ecol. 2017 Nov;26(21):5872-5895. doi: 10.1111/mec.14350. Epub 2017 Oct 26. Mol Ecol. 2017. PMID: 28921802 Review.
References
Associated data
LinkOut - more resources
Full Text Sources

