Inhibition of PHLDA3 expression in human superoxide dismutase 1-mutant amyotrophic lateral sclerosis astrocytes protects against neurotoxicity
- PMID: 39144751
- PMCID: PMC11323778
- DOI: 10.1093/braincomms/fcae244
Inhibition of PHLDA3 expression in human superoxide dismutase 1-mutant amyotrophic lateral sclerosis astrocytes protects against neurotoxicity
Abstract
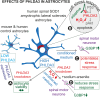
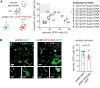
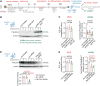
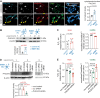
Pleckstrin homology-like domain family A-member 3 (PHLDA3) has recently been identified as a player in adaptive and maladaptive cellular stress pathways. The outcome of pleckstrin homology-like domain family A-member 3 signalling was shown to vary across different cell types and states. It emerges that its expression and protein level are highly increased in amyotrophic lateral sclerosis (ALS) patient-derived astrocytes. Whether it orchestrates a supportive or detrimental function remains unexplored in the context of neurodegenerative pathologies. To directly address the role of pleckstrin homology-like domain family A-member 3 in healthy and ALS astrocytes, we used overexpression and knockdown strategies. We generated cultures of primary mouse astrocytes and also human astrocytes from control and ALS patient-derived induced pluripotent stem cells harbouring the superoxide dismutase 1 mutation. Then, we assessed astrocyte viability and the impact of their secretome on oxidative stress responses in human stem cell-derived cortical and spinal neuronal cultures. Here, we show that PHLDA3 overexpression or knockdown in control astrocytes does not significantly affect astrocyte viability or reactive oxygen species production. However, PHLDA3 knockdown in ALS astrocytes diminishes reactive oxygen species concentrations in their supernatants, indicating that pleckstrin homology-like domain family A-member 3 can facilitate stress responses in cells with altered homeostasis. In support, supernatants of PHLDA3-silenced ALS and even control spinal astrocytes with a lower pleckstrin homology-like domain family A-member 3 protein content could prevent sodium arsenite-induced stress granule formation in spinal neurons. Our findings provide evidence that reducing pleckstrin homology-like domain family A-member 3 levels may transform astrocytes into a more neurosupportive state relevant to targeting non-cell autonomous ALS pathology.
Keywords: PHLDA3; amyotrophic lateral sclerosis; astrocyte; astrocyte–neuron interaction; cell stress.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
A.L. is a scientific consultant to Tachyon Ventures, a venture capital firm (Los Angeles, CA, USA). However, the consultancy is not pertinent to the work presented in this manuscript. The rest of the authors declare no conflict of interest.
Figures




References
-
- Vahsen BF, Gray E, Thompson AG, et al. Non-neuronal cells in amyotrophic lateral sclerosis—From pathogenesis to biomarkers. Nat Rev Neurol. 2021;17(6):333–348. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
