Prediction and verification of benignancy and malignancy of pulmonary nodules based on inflammatory related biological markers
- PMID: 39144966
- PMCID: PMC11320450
- DOI: 10.1016/j.heliyon.2024.e34585
Prediction and verification of benignancy and malignancy of pulmonary nodules based on inflammatory related biological markers
Abstract
Objective: Inflammation plays an important role in the transformation of pulmonary nodules (PNs) from benign to malignant. Prediction of benignancy and malignancy of PNs is still lacking efficacy methods. Although Mayo or Brock model have been widely applied in clinical practices, their application conditions are limited. This study aims to construct a diagnostic model of PNs by machine learning using inflammation-related biological markers (IRBMs).
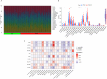
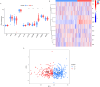
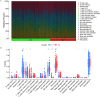
Methods: Inflammatory related genes (IRGs) were first extracted from GSE135304 chip data. Then, differentially expressed genes (DEGs) and infiltrating immune cells were screened between malignant pulmonary nodules (MN) and benign pulmonary nodule (BN). Correlation analysis was performed on DEGs and infiltrating immune cells. Molecular modules of IRGs were identified through Consistency cluster analysis. Subsequently, IRBMs in IRGs modules were filtered through Weighted gene co-expression network analysis (WGCNA). An optimal diagnostic model was established using machine learning methods. Finally, external dataset GSE108375 was used to verify this result.
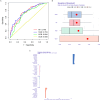
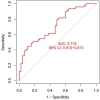
Results: 4 hub IRGs and 3 immune cells showed significantly difference between MN and BN, C1 and C2 module, namely PRTN3, ELANE, NFKB1 and CTLA4, T cells CD4 naïve, NK cells activated and Monocytes. IRBMs were screened from black module and yellowgreen module through WGCNA analysis. The Support vector machines (SVM) was identified as the optimal model with the Area Under Curve (AUC) was 0.753. A nomogram was established based on 5 hub IRBMs, namely HS.137078, KLC3, C13ORF15, STOM and KCTD13. Finally, external dataset GSE108375 verified this result, with the AUC was 0.718.
Conclusion: SVM model established by 5 hub IRBMs was able to effectively identify MN or BN. Accumulating inflammation and immune dysfunction were important to the transformation from BN to MN.
Keywords: Immune cells infiltration; Inflammation-related biological markers; Nomogram; Pulmonary nodules; Support vector machine.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

