Pan-cancer bioinformatics analysis of hepatic leukemia factor and further validation in colorectal cancer
- PMID: 39145052
- PMCID: PMC11319992
- DOI: 10.21037/tcr-23-2274
Pan-cancer bioinformatics analysis of hepatic leukemia factor and further validation in colorectal cancer
Abstract
Background: Hepatic leukemia factor (HLF) is associated with cancer onset, growth, and progression; however, little is known regarding its biological role in pan-cancer. In order to further evaluate the diagnostic and prognostic value of HLF in pan-cancer and colorectal cancer (CRC), we performed comprehensive bioinformatics analyses of the molecular mechanism of HLF in pan-cancer, with subsequent verification in CRC.
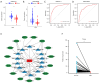
Methods: We downloaded data (gene expression, clinical data, follow-up duration, and immune-related data) related to 33 solid tumor types from UCSC Xena (University of California Santa Cruz cancer database, https://xena.ucsc.edu/). HLF expression was analyzed in pan-cancer, and its diagnostic efficacy, prognostic value, and correlation with pathological stage and cancer immunity were determined. We also analyzed gene alterations in HLF and biological processes involved in its regulation in pan-cancer. Using CRC data in The Cancer Genome Atlas (TCGA), we assessed correlations between HLF and CRC diagnosis, prognosis, and drug sensitivity and performed functional enrichment analyses. Moreover, we constructed an HLF-related ceRNA regulatory network. Finally, we externally validated HLF expression and diagnostic and prognostic value in CRC using Gene Expression Omnibus (GEO) database, as well as by performing in vitro experiments.
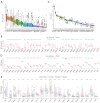
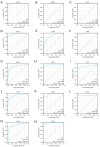
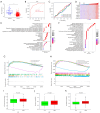
Results: HLF expression was downregulated in most tumors, and HLF showed good predictive potential for pan-cancer diagnosis and prognosis. It was closely related to the clinicopathological stages of pan-cancer. Further, HLF was associated with tumor microenvironment and immune cell infiltration in many tumors. Analyses involving cBioPortal revealed changes in HLF amplifications and mutations in most tumors. HLF was also closely associated with microsatellite instability and tumor mutational burden in pan-cancer and involved in regulating various tumor-related pathways and biological processes. In CRC, HLF expression was similarly downregulated, with implications for CRC diagnosis and prognosis. Functional enrichment analysis indicated the association of HLF with many cancer-related pathways. Further, HLF was associated with drug (e.g., oxaliplatin) sensitivity in CRC. The ceRNA regulatory network showed multigene regulation of HLF in CRC. External validation involving GEO databases and quantitative real-time polymerase chain reaction (qRT-PCR) data substantiated these findings.
Conclusions: HLF expression generally exhibited downregulation in pan-cancer, contributing to tumor occurrence and development by regulating various biological processes and affecting tumor immune characteristics. HLF was also closely related to CRC occurrence and development. We believe HLF can serve as a reliable diagnostic, prognostic, and immune biomarker for pan-cancer.
Keywords: Hepatic leukemia factor (HLF); cancer immunity; diagnosis; pan-cancer; prognosis.
2024 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2274/coif). The authors have no conflicts of interest to declare.
Figures





References
LinkOut - more resources
Full Text Sources
